Information
Documentation
cell_explorer: Unit Cell Explorer
Synopsis
cell_explorer indexing-results.stream
Description
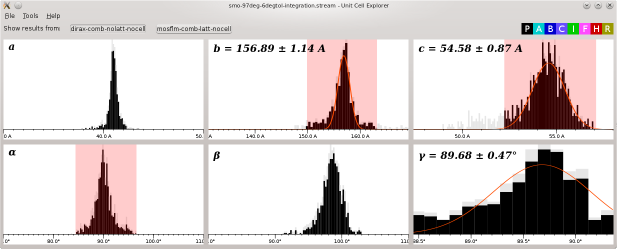
The Unit Cell Explorer allows you to visualise the distribuftions of unit cell parameters resulting from processing a series of diffraction patterns with indexamajig.
Start the Cell Explorer simply by executing cell_explorer followed by the filename of the stream which contains your indexing results.
The Cell Explorer window shows histograms for, left to right respectively, a, b and c in the top row and alpha, beta and gamma on the bottom row.
Click and drag to move the graphs to the left and right. Scroll up and down to zoom in and out. Press plus or minus to increase or decrease the number of bins used to calculate the histogram. Changing the binning is usually necessary when zooming in or out by a large amount.
Different colours are used to represent different centering types (P, A, B, C, I, F, H and R) according to the colour key at the top right. Click one of the squares in the key to change the colour used for that centering type to grey, and again to change it to black.
Click and drag with shift held down to select a range of values in one of the histograms. Unit cells which have that parameter outside the selected range will be shown in grey in all the histograms.
After selecting a range of values, use Tools->Fit Cell (or press Ctrl+F) to fit a normal distribution to the histogram data. The mean and standard deviation of the parameter will be shown. Unit cells will not be included in the fit if they are outside the ranges selected for the other parameters.
The indexing algorithms found in the stream are shown as buttons at the top. Click one of the buttons to deselect it, and again to select it again. Unit cells from deselected algorithms will not be shown in the histograms (not even in grey)
Author
This page was written by Thomas White.
Reporting bugs
Report bugs to taw@physics.org
Copyright and Disclaimer
Copyright © 2014-2020 Deutsches Elektronen-Synchrotron DESY, a research centre of the Helmholtz Association.
Please read the AUTHORS file in the CrystFEL source code distribution for a full list of contributions and contributors.
CrystFEL is free software: you can redistribute it and/or modify it under the terms of the GNU General Public License as published by the Free Software Foundation, either version 3 of the License, or (at your option) any later version.
CrystFEL is distributed in the hope that it will be useful, but WITHOUT ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the GNU General Public License for more details.
You should have received a copy of the GNU General Public License along with CrystFEL. If not, see http://www.gnu.org/licenses/.
See also
| crystfel | indexamajig |
If CrystFEL is installed on your computer, you can read this manual page offline using the command man cell_explorer.


