Information
Documentation
Examples of research using CrystFEL
This page contains references to research for which CrystFEL was used. We track PDB entries and use citation alerts to keep this list up to date, but occasionally things get missed out. If you've published a paper about research for which CrystFEL was important, and it doesn't already appear here, please get in touch so we can include your article.
Note: articles are grouped by year according to their date of first publication, whereas the citations show the publication date of the journal issue. There can be differences between these years when articles are published near the end of the year.
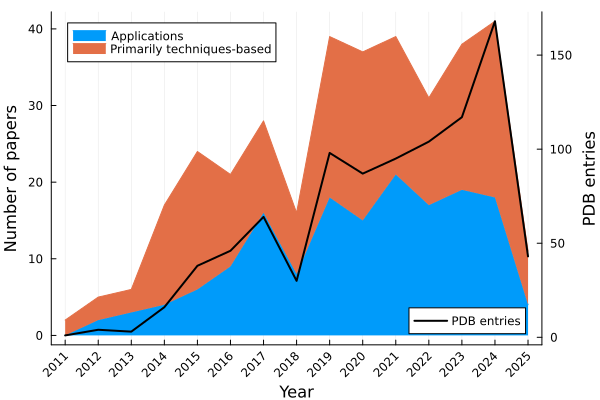
At the time of writing, CrystFEL has been used for 383 total papers and 1097 PDB entries.
2026 (5 articles and 30 PDB entries this year)
- Diandra Doppler, Alice Grieco, Domin Koh, Abhik Manna, et al. "Minimized sample consumption for time-resolved serial crystallography applied to the redox cycle of human NQO1". Communications Chemistry (2026). doi:10.1038/s42004-026-01908-9 (open access). PDB IDs 9ezt, 9ezr, 9id0, 9ezs and 9ezq.
- Jungmin Kang, Yoshiaki Shimazu, Fangjia Luo, Ayumi Yamashita, et al. "Compact tape-driven sample delivery system for serial femtosecond crystallography". Journal of Applied Crystallography 59 (2026). doi:10.1107/s1600576726000063 (open access - download PDF). PDB IDs 9v3d, 9v3g, 9v3h, 9v3i, 9v3j, 9v3k, 9v3l, 9v3m, 9v3n, 9v3o, 9v3p, 9v3q and 9v3r.
- Ronald Rios-Santacruz, Harshwardhan Poddar, Kevin Pounot, Derren J. Heyes, et al. "Integrated structural dynamics uncover a new B12 photoreceptor activation mode". Nature (2026). doi:10.1038/s41586-025-10074-2 PDB IDs 9s06, 9s07, 9s08, 9s09, 9s0a, 9s0b, 9s0c, 9s0d, 9s0e, 9s0f and 9s0j.
- Ki Hyun Nam and Sang-Ho Na. "Serial femtosecond crystallography data processing at the global science data hub center at KISTI". Scientific Reports (2026). doi:10.1038/s41598-026-36540-z (open access).
- Alessia Pepe, Ronald Rios-Santacruz, Sara Schianchi, Jovana Vitas, et al. "Guanosine hydrogel as a new injection matrix for protein serial X-ray crystallography". Journal of Applied Crystallography 59 (2026) 135-145. doi:10.1107/s1600576725011276 PDB ID 9qtl.
2025 (33 articles and 175 PDB entries this year)
- Gargi Gore, Andreas Prester, David von Stetten, Kim Bartels, et al. "Binding mode of Isoxazolyl Penicillins to a Class-A β-lactamase at ambient conditions". Communications Chemistry 8 (2025). doi:10.1038/s42004-025-01801-x (open access - download PDF). PDB IDs 9r0r, 9rao and 9raj.
- Takeshi Murakawa, Mamoru Suzuki, Kenji Fukui, Tetsuya Masuda, et al. "Real-time capture of domain movements during copper amine oxidase catalysis by mix-and-inject serial crystallography". Nature Communications 16 (2025). doi:10.1038/s41467-025-67230-5 (open access - download PDF). PDB IDs 9lz3, 9lz4, 9lz5, 9lz6, 9lz7, 9lz8, 9lz9, 9lza, 9lzb, 9lzc, 9lzd, 9lze and 9lzf.
- Sreelaja Pulleri Vadhyar, Ehsan Nikbin, Hazem Daoud, Jane Y. Howe, et al. "Room-temperature structure determination of vacuum-sensitive organic compounds by formvar encapsulation and serial electron diffraction". Journal of Applied Crystallography 58 (2025) 2119-2124. doi:10.1107/s1600576725009823 (open access - download PDF). CCDC IDs 2464360 and 2479753.
- Jaehyun Park, Sehan Park and Ki Hyun Nam. "Preliminary Serial Femtosecond Crystallography Studies of Myoglobin from Equine Skeletal Muscle". Crystals 15 (2025) 905. doi:10.3390/cryst15100905 (open access - download PDF). PDB ID 9x07.
- Dmitrii Zabelskii, Ekaterina Round, Huijong Han, David von Stetten, et al. "Viscoelastic characterization of the lipid cubic phase provides insights into high-viscosity extrusion injection for XFEL experiments". Scientific Reports 15 (2025). doi:10.1038/s41598-025-25449-8 (open access - download PDF). PDB IDs 9i6h, 9i6i, 9i6j, 9i6k, 9i6l, 9i6m, 9i6n, 9i6o and 9i6p.
- Alaleh Shafiei, Nilufer Baldir, Jongbum Na, Jin Hae Kim, et al. "Comparative Structural Analysis of Escherichia Coli Cyay at Room and Cryogenic Temperatures Using Macromolecular and Serial Crystallography". ChemBioChem 26 (2025). doi:10.1002/cbic.202500442 (open access - download PDF). PDB ID 9v63.
- Madan Kumar Shankar, Lukas Grunewald, Weixiao Yuan Wahlgren, Brigitte Stucki-Buchli, et al. "Ultrafast, remote-controlled protonation reaction enables structural changes in a phytochrome". Science Advances 11 (2025). doi:10.1126/sciadv.ady0499 PDB IDs 8c3i, 7gqv, 7gqw, 7gqx, 7gqy, 7gqz, 7gr0, 7gr1, 7gr2, 7gr3, 7gr4, 7gr5, 7gr6, 7gr7, 7gr8, 7gr9, 7gra, 7grb, 7grc and 7grd.
- Sebastian Günther, Pontus Fischer, Marina Galchenkova, Sven Falke, et al. "Room-temperature X-ray fragment screening with serial crystallography". Nature Communications 16 (2025). doi:10.1038/s41467-025-64918-6 (open access - download PDF). PDB IDs 9s5b, 9g1a, 9g1b, 9g1c, 9g1d, 9g1e, 9g1f, 9g1g and 9g1h.
- Alexander Gorel, Robert L. Shoeman, Elisabeth Hartmann, Stanislaw Nizinski, et al. "Testing the limits: serial crystallography using unpatterned fixed targets". IUCrJ 12 (2025). doi:10.1107/s2052252525008371 (open access - download PDF). PDB IDs 9gt0, 9gt1, 9gt2, 9gt3, 9gt4, 9gt5, 9gt6, 9gt7, 9gt8, 9gt9 and 9gta.
- Esra Ayan, Sylvain Engilberge, Shun Yokoi, Julien Orlans, et al. "Triple Calcium Binding Stoichiometry in the Monoclinic Crystal Form of Protracted Insulin". Small Structures (2025). doi:10.1002/sstr.202500398 (open access - download PDF). PDB ID 9utk.
- Peter Smyth, Sofia Jaho, Lewis J. Williams, Gabriel Karras, et al. "Time-resolved serial synchrotron and serial femtosecond crystallography of heme proteins using photocaged nitric oxide". IUCrJ 12 (2025) 582-594. doi:10.1107/s2052252525006645 (open access - download PDF). PDB ID 9hqt.
- Niko W. Vlahakis, Cameron W. Flowers, Mengting Liu, Matthew P. Agdanowski, et al. "Combining MicroED and native mass spectrometry for structural discovery of enzyme–small molecule complexes". Proceedings of the National Academy of Sciences 122 (2025). doi:10.1073/pnas.2503780122 (open access - download PDF). PDB IDs 9nbf, 9nbj, 9nbk, 9nbn, 9nb4, 9nb7 and 9ncc.
- Eike C. Schulz, Andreas Prester, David von Stetten, Gargi Gore, et al. "Probing the modulation of enzyme kinetics by multi-temperature, time-resolved serial crystallography". Nature Communications 16 (2025). doi:10.1038/s41467-025-61631-2 (open access - download PDF). PDB IDs 9g81, 9g7y, 9g82, 9g7z, 9g7x, 9g6n, 9g80, 9g6l, 9g61, 9g7v, 9g6o, 9g7w, 9g6p, 9g6m, 9g5x, 9g5w, 9i7l and 9g5s.
- Sergi Plana-Ruiz, Penghan Lu, Govind Ummethala and Rafal E. Dunin-Borkowski. "On the use of beam precession for serial electron crystallography". Journal of Applied Crystallography 58 (2025). doi:10.1107/s1600576725005606 (open access - download PDF).
- Soshichiro Nagano, David von Stetten, Kaoling Guan, Peng-Yuan Chen, et al. "Pr and Pfr structures of plant phytochrome A". Nature Communications 16 (2025). doi:10.1038/s41467-025-60738-w (open access - download PDF). PDB IDs 9f4i, 9er4 and 9qzt. CXIDB ID 224.
- Ki Hyun Nam. "Effect of crystal-to-detector distance shift on data processing in serial crystallography". PLOS One 20 (2025) e0327019. doi:10.1371/journal.pone.0327019 (open access). PDB ID 9ugi.
- Tek Narsingh Malla, Luis Aldama, Viridiana Leon, Denisse Feliz, et al. "Observation of early events in the photoactivation of Myxobacterial phytochrome using time-resolved serial femtosecond crystallography". Communications Chemistry 8 (2025). doi:10.1038/s42004-025-01578-z (open access - download PDF). PDB IDs 9dz3, 9md9, 9meb and 9dzp. CXIDB ID 234.
- Manashi Sonowal, Gihan Ketawala, Nirupa Nagaratnam, Dhenugen Logeswaran, et al. "Functional implications of hexameric dynamics in
SARS ‐CoV ‐2 Nsp15". Protein Science 34 (2025). doi:10.1002/pro.70115 (open access - download PDF). PDB IDs 9mru, 9mry and 9mrw. - Ebru Destan, Engin Turkut, Alper Aldeniz, Jungmin Kang, et al. "Experimental and Computational Insights into the Structural Dynamics of the Fc Fragment of IgG1 Subtype from Biosimilar VEGF‐Trap". Small Structures (2025). doi:10.1002/sstr.202400680 (open access - download PDF). PDB ID 8zck.
- Manuel Maestre-Reyna, Yuhei Hosokawa, Po-Hsun Wang, Martin Saft, et al. "Capturing structural intermediates in an animal-like cryptochrome photoreceptor by time-resolved crystallography". Science Advances 11 (2025). doi:10.1126/sciadv.adu7247 PDB IDs 8z1j, 8z24, 8z26, 8z2d, 8z44, 8z3d, 8z3g, 8z3l, 8z3x, 8z41, 8z45, 8z4k, 8z4m, 8z4p, 8z4u, 8z6i, 8z6j, 8z6k, 8z8f, 8z8k, 8za8 and 8zlr.
- Ban H. Al-Tayyem, Philipp Müscher-Polzin, Kanupriya Pande, Oleksandr Yefanov, et al. "In situ monitoring of ligand-to-metal energy transfer in combination with synchrotron-based X-ray diffraction methods to elucidate the synthesis mechanism and structural evolution of lanthanide complexes". Frontiers in Chemistry 13 (2025). doi:10.3389/fchem.2025.1536383 (open access). CCDC ID 2427847.
- Po-Hsun Wang, Yuhei Hosokawa, Jessica C Soares, Hans-Joachim Emmerich, et al. "Redox-State-Dependent Structural Changes within a Prokaryotic 6–4 Photolyase". Journal of the American Chemical Society (2025). doi:10.1021/jacs.4c18116 (open access - download PDF). PDB IDs 9hno, 9hnn and 9hnm.
- Lewis J. Williams, Amy J. Thompson, Philipp Dijkstal, Martin Appleby, et al. "Damage before destruction? X-ray-induced changes in single-pulse serial femtosecond crystallography". IUCrJ 12 (2025) 358-371. doi:10.1107/s2052252525002660 (open access - download PDF). PDB IDs 9eqz, 9er0, 9equ, 9eqs, 9er1, 9eqt, 9eqr, 9eqx, 9eqy, 9eqv, 9epd, 9epg, 9epk, 9epj, 9epe, 9epi and 9eph.
- Robert Bosman, Giorgia Ortolani, Swagatha Ghosh, Daniel James, et al. "Structural basis for the prolonged photocycle of sensory rhodopsin II revealed by serial synchrotron crystallography". Nature Communications 16 (2025). doi:10.1038/s41467-025-58263-x (open access - download PDF). PDB IDs 9h20, 9h1x, 8pwp, 8pwj, 8pwi, 8pwg, 8pwq and 9h1w.
- Owens Uwangue, Johan Glerup, Andreas Dunge, Monika Bjelcic, et al. "Microcrystallization and room-temperature serial crystallography structure of human cytochrome P450 3A4". Archives of Biochemistry and Biophysics (2025) 110419. doi:10.1016/j.abb.2025.110419 (open access). PDB ID 9gk1.
- Satoshi Nagao, Wako Kuwano, Takehiko Tosha, Keitaro Yamashita, et al. "XFEL crystallography reveals catalytic cycle dynamics during non-native substrate oxidation by cytochrome P450BM3". Communications Chemistry 8 (2025). doi:10.1038/s42004-025-01440-2 (open access - download PDF). PDB IDs 8yay, 8yaz, 8yb0, 8yb1, 8yb2 and 8yb3.
- Ki Hyun Nam. "Effects of Beam Center Position Shifts on Data Processing in Serial Crystallography". Crystals 15 (2025) 185. doi:10.3390/cryst15020185 (open access - download PDF). PDB IDs 9lmk and 9lml.
- Do-Heon Gu, Dong Tak Jeong, Cheolsoo Eo, Pil-Won Seo, et al. "Novel fixed-target serial crystallography flip-holder for macromolecular crystallography beamlines at synchrotron radiation sources". Journal of Synchrotron Radiation 32 (2025). doi:10.1107/s1600577524011664 (open access - download PDF).
- Jérôme Kieffer, Julien Orlans, Nicolas Coquelle, Samuel Debionne, et al. "Application of signal separation to diffraction image compression and serial crystallography". Journal of Applied Crystallography 58 (2025) 138-153. doi:10.1107/s1600576724011038 (open access - download PDF).
- Justin Trujillo, Russell Fung, Madan Kumar Shankar, Peter Schwander, et al. "Filling data analysis gaps in time-resolved crystallography by machine learning". Structural Dynamics 12 (2025). doi:10.1063/4.0000280 (open access - download PDF).
- Ki Hyun Nam. "Impact of Diffraction Data Volume on Data Quality in Serial Crystallography". Crystals 15 (2025) 104. doi:10.3390/cryst15020104 (open access - download PDF).
- Ki Hyun Nam. "Temperature-Dependent Structural Changes of the Active Site and Substrate-Binding Cleft in Hen Egg White Lysozyme". Crystals 15 (2025) 111. doi:10.3390/cryst15020111 (open access - download PDF). PDB ID 8ybg.
- Julien Orlans, Samuel L. Rose, Gavin Ferguson, Marcus Oscarsson, et al. "Advancing macromolecular structure determination with microsecond X-ray pulses at a 4th generation synchrotron". Communications Chemistry 8 (2025). doi:10.1038/s42004-024-01404-y (open access - download PDF). PDB IDs 9ftu, 9fts, 9ftv, 9ftx, 9fty, 9fu1, 9fud, 9fue and 9fup.
2024 (42 articles and 182 PDB entries this year)
- Kara A. Zielinski, Cole Dolamore, Kevin M. Dalton, Nathan Smith, et al. "Resolving DJ-1 Glyoxalase Catalysis Using Mix-and-Inject Serial Crystallography at a Synchrotron". Pre-print (2024). doi:10.1101/2024.07.19.604369 PDB IDs 9cge, 9cgg, 9cgf, 9cfq, 9cgd, 9cga and 9cgb.
- Matthias Mulder, Songhwan Hwang, Matthias Broser, Steffen Brünle, et al. "Structural Insights Into the Opening Mechanism of C1C2 Channelrhodopsin". Journal of the American Chemical Society 147 (2024) 1282-1290. doi:10.1021/jacs.4c15402 (open access - download PDF). PDB IDs 9go1 and 9go2.
- Hannah Glover, Torben Saßmannshausen, Quentin Bertrand, Matilde Trabuco, et al. "Photoswitch dissociation from a G protein-coupled receptor resolved by time-resolved serial crystallography". Nature Communications 15 (2024). doi:10.1038/s41467-024-55109-w (open access - download PDF). PDB IDs 8rwh, 8rwi, 8rvw and 8rwx.
- Thomas White, Tim Schoof, Sergey Yakubov, Aleksandra Tolstikova, et al. "Real-time data processing for serial crystallography experiments". IUCrJ 12 (2025) 97-108. doi:10.1107/s2052252524011837 (open access - download PDF). PDB ID 8rpm.
- Jihan Kim, Jaehyun Park, Keondo Lee, Wan Kyun Chung, et al. "Exploring the reaction dynamics of alanine racemase using serial femtosecond crystallography". Scientific Reports 14 (2024). doi:10.1038/s41598-024-83045-8 (open access - download PDF). PDB IDs 8zpe, 8zpf, 8zpg, 8zph and 9jt7.
- Abhik Manna, Mukul Sonker, Domin Koh, Michael Steiger, et al. "Cyclic Olefin Copolymer-Based Fixed-Target Sample Delivery Device for Protein X-ray Crystallography". Analytical Chemistry (2024). doi:10.1021/acs.analchem.4c03484 (open access - download PDF). PDB ID 9fa5.
- R. Bruce Doak, Robert L. Shoeman, Alexander Gorel, Stanisław Niziński, et al. "Sheet-on-sheet fixed target data collection devices for serial crystallography at synchrotron and XFEL sources". Journal of Applied Crystallography 57 (2024) 1725-1732. doi:10.1107/s1600576724008914 (open access - download PDF). PDB ID 9g2j.
- Quentin Bertrand, Przemyslaw Nogly, Eriko Nango, Demet Kekilli, et al. "Structural effects of high laser power densities on an early bacteriorhodopsin photocycle intermediate". Nature Communications 15 (2024). doi:10.1038/s41467-024-54422-8 (open access - download PDF). PDB IDs 9f9b, 9f9c, 9f9d, 9f9e, 9f9f, 9f9g, 9f9h, 9f9i and 9f9j.
- Tek Narsingh Malla, Srinivasan Muniyappan, David Menendez, Favour Ogukwe, et al. "Exploiting fourth-generation synchrotron radiation for enzyme and photoreceptor characterization". IUCrJ 12 (2025). doi:10.1107/s2052252524010868 (open access - download PDF). PDB IDs 9cuf, 9d10 and 9d2h.
- Andrea Moreno, Isabel Quereda-Moraleda, Celia Lozano-Vallhonrat, María Buñuel-Escudero, et al. "New insights into the function and molecular mechanisms of Ferredoxin-NADP+ reductase from Brucella ovis". Archives of Biochemistry and Biophysics (2024) 110204. doi:10.1016/j.abb.2024.110204 (open access). PDB IDs 9gxc and 9gxb.
- Jaewon Yang, Hyung Jin Jeon, Seonha Park, Junga Park, et al. "Structural Insights and Catalytic Mechanism of 3-Hydroxybutyryl-CoA Dehydrogenase from Faecalibacterium Prausnitzii A2-165". International Journal of Molecular Sciences 25 (2024) 10711. doi:10.3390/ijms251910711 (open access - download PDF). PDB ID 9jhy.
- Sofia M. Kapetanaki, Nicolas Coquelle, David von Stetten, Martin Byrdin, et al. "Crystal structure of a bacterial photoactivated adenylate cyclase determined by serial femtosecond and serial synchrotron crystallography". IUCrJ 11 (2024) 991-1006. doi:10.1107/s2052252524010170 (open access - download PDF). PDB IDs 9f1w and 9f1x.
- Elke De Zitter, David Perl, Martin Savko, Daniel W. Paley, et al. "Elucidating metal–organic framework structures using synchrotron serial crystallography". CrystEngComm 26 (2024) 5644-5654. doi:10.1039/d4ce00735b (open access - download PDF). CCDC IDs 2353477 and 2353480.
- A. Dunge, C. Phan, O. Uwangue, M. Bjelcic, et al. "Exploring serial crystallography for drug discovery". IUCrJ 11 (2024) 831-842. doi:10.1107/s2052252524006134 (open access - download PDF). PDB IDs 8qwg, 8qvf, 8qwi, 8qvh, 8qvk, 8qvl, 8qvm and 8qvg.
- Basudev Maity, Mitsuo Shoji, Fangjia Luo, Takanori Nakane, et al. "Real-time observation of a metal complex-driven reaction intermediate using a porous protein crystal and serial femtosecond crystallography". Nature Communications 15 (2024). doi:10.1038/s41467-024-49814-9 (open access - download PDF). PDB IDs 8wzt, 8wzf, 8wzg, 8wzr and 8wzv. CXIDB ID 221.
- Nicolas Foos, Jean-Baptise Florial, Mathias Eymery, Jeremy Sinoir, et al. "In situ serial crystallography facilitates 96-well plate structural analysis at low symmetry". IUCrJ 11 (2024) 780-791. doi:10.1107/s2052252524005785 (open access - download PDF). PDB IDs 9eu5 and 9ent.
- Adams Vallejos, Gergely Katona and Richard Neutze. "Appraising protein conformational changes by resampling time-resolved serial x-ray crystallography data". Structural Dynamics 11 (2024). doi:10.1063/4.0000258
- Guillaume Gotthard, Sandra Mous, Tobias Weinert, Raiza Nara Antonelli Maia, et al. "Capturing the blue-light activated state of the Phot-LOV1 domain from Chlamydomonas reinhardtii using time-resolved serial synchrotron crystallography". IUCrJ 11 (2024) 792-808. doi:10.1107/s2052252524005608 (open access - download PDF). PDB IDs 8qi9, 8qil, 8qiq, 8qib, 8qit, 8qiw, 8qip, 8qig, 8qim, 8qin, 8qis, 8qiv, 8qih, 8qio, 8qia, 8qii, 8qir, 8qik, 8qiu and 8qif.
- Andreas Prester, Markus Perbandt, Marina Galchenkova, Dominik Oberthuer, et al. "Time-resolved crystallography of boric acid binding to the active site serine of the β-lactamase CTX-M-14 and subsequent 1,2-diol esterification". Communications Chemistry 7 (2024). doi:10.1038/s42004-024-01236-w (open access - download PDF). PDB IDs 8pcv, 8pcl, 8pcq, 8pc9, 8pcc, 8pci, 8pcj, 8pcs, 8pcg, 8pcm, 8pct, 8pcp, 8pca, 8pcb, 8pcf, 8pcn, 8pco, 8pck, 8pcr, 8pce, 8pcu and 8pcd.
- Guillaume Gotthard, Andrea Flores-Ibarra, Melissa Carrillo, Michal W. Kepa, et al. "Fixed-target pump–probe SFX: eliminating the scourge of light contamination". IUCrJ 11 (2024) 749-761. doi:10.1107/s2052252524005591 (open access - download PDF).
- Piero Gasparotto, Luis Barba, Hans-Christian Stadler, Greta Assmann, et al. "TORO Indexer: a PyTorch-based indexing algorithm for kilohertz serial crystallography". Journal of Applied Crystallography 57 (2024) 931-944. doi:10.1107/s1600576724003182 (open access - download PDF).
- James M. Baxter, Christopher D. M. Hutchison, Alisia Fadini, Karim Maghlaoui, et al. "Power Density Titration of Reversible Photoisomerization of a Fluorescent Protein Chromophore in the Presence of Thermally Driven Barrier Crossing Shown by Quantitative Millisecond Serial Synchrotron X-ray Crystallography". Journal of the American Chemical Society 146 (2024) 16394-16403. doi:10.1021/jacs.3c12883 (open access - download PDF). PDB IDs 8ul0, 8ul1, 8ul2, 8ul3, 8ul4 and 8ul5.
- Patrick Adams, Tamar L. Greaves and Andrew V. Martin. "Crystal structure via fluctuation scattering". IUCrJ 11 (2024) 538-555. doi:10.1107/s2052252524003932 (open access - download PDF).
- Monika Bjelčić, Oskar Aurelius, Jie Nan, Richard Neutze, et al. "Room-temperature serial synchrotron crystallography structure of Spinacia oleracea RuBisCO". Acta Crystallographica Section F Structural Biology Communications 80 (2024) 117-124. doi:10.1107/s2053230x24004643 (open access - download PDF). PDB ID 8qj0.
- Robert Bosman, Andreas Prester, Sihyun Sung, Lea von Soosten, et al. "A systematic comparison of Kapton-based HARE chips for fixed-target serial crystallography". Cell Reports Physical Science 5 (2024) 101987. doi:10.1016/j.xcrp.2024.101987 (open access). PDB IDs 8reg, 8rsh, 8rse, 8rei, 8reh, 8rsx and 8rs2.
- Alice Grieco, Sergio Boneta, José A. Gavira, Angel L. Pey, et al. "Structural dynamics and functional cooperativity of human
NQO1 by ambient temperature serial crystallography and simulations". Protein Science 33 (2024). doi:10.1002/pro.4957 (open access - download PDF). PDB IDs 8rfn and 8rfm. - Jake A. Hill, Yvonne Nyathi, Sam Horrell, David von Stetten, et al. "An ultraviolet-driven rescue pathway for oxidative stress to eye lens protein human gamma-D crystallin". Communications Chemistry 7 (2024). doi:10.1038/s42004-024-01163-w (open access - download PDF). PDB IDs 8q3l, 8bd0 and 8bpi.
- Andrea Cellini, Madan Kumar Shankar, Amke Nimmrich, Leigh Anna Hunt, et al. "Directed ultrafast conformational changes accompany electron transfer in a photolyase as resolved by serial crystallography". Nature Chemistry 16 (2024) 624-632. doi:10.1038/s41557-023-01413-9 (open access - download PDF). PDB IDs 8c1u, 8c6c, 8c69, 8c6f, 8c6b, 8c6h and 8c6a. CXIDB ID 219.
- Patrick Y. A. Reinke, Robin Schubert, Dominik Oberthür, Marina Galchenkova, et al. "SARS-CoV-2 Mpro responds to oxidation by forming disulfide and NOS/SONOS bonds". Nature Communications 15 (2024). doi:10.1038/s41467-024-48109-3 (open access - download PDF). PDB IDs 7pxz and 7pzq.
- Samuel Perrett, Alisia Fadini, Christopher D. M. Hutchison, Sayantan Bhattacharya, et al. "Kilohertz droplet-on-demand serial femtosecond crystallography at the European XFEL station FXE". Structural Dynamics 11 (2024). doi:10.1063/4.0000248 PDB ID 8rus.
- Kiyofumi Takaba, Saori Maki-Yonekura, Ichiro Inoue, Kensuke Tono, et al. "Comprehensive Application of XFEL Microcrystallography for Challenging Targets in Various Organic Compounds". Journal of the American Chemical Society 146 (2024) 5872-5882. doi:10.1021/jacs.3c11523 CXIDB IDs 223 and 223. CCDC IDs 2296549, 2296550, 2296551 and 2270804.
- Jihan Kim, Sehan Park, Yunje Cho and Jaehyun Park. "Application of Micro-Tubing Reeling System to Serial Femtosecond Crystallography". Photonics 11 (2024) 95. doi:10.3390/photonics11010095 (open access - download PDF). PDB ID 8jqv.
- Jack Stubbs, Theo Hornsey, Niall Hanrahan, Luis Blay Esteban, et al. "Droplet microfluidics for time-resolved serial crystallography". IUCrJ 11 (2024) 237-248. doi:10.1107/s2052252524001799 (open access - download PDF). PDB IDs 8s2v, 8s2u, 8s2w and 8s2x.
- Joshua A. Hull, Cheol Lee, Jin Kyun Kim, Seon Woo Lim, et al. "XFEL structure of carbonic anhydrase II: a comparative study of XFEL, NMR, X-ray and neutron structures". Acta Crystallographica Section D Structural Biology 80 (2024) 194-202. doi:10.1107/s2059798324000482 PDB ID 8sf1.
- Robert Schönherr, Juliane Boger, J. Mia Lahey-Rudolph, Mareike Harms, et al. "A streamlined approach to structure elucidation using in cellulo crystallized recombinant proteins, InCellCryst". Nature Communications 15 (2024). doi:10.1038/s41467-024-45985-7 (open access - download PDF). PDB IDs 8c51, 8c53, 8c5k, 8cd4, 8cd5 and 8cd6.
- Thomas R. M. Barends, Alexander Gorel, Swarnendu Bhattacharyya, Giorgio Schirò, et al. "Influence of pump laser fluence on ultrafast myoglobin structural dynamics". Nature 626 (2024) 905-911. doi:10.1038/s41586-024-07032-9 (open access - download PDF). PDB IDs 8r8f, 8r8g, 8r8h, 8r8i, 8r8j, 8r8w, 8r8x, 8r8y, 8r8z, 8r90, 8r91, 8r92, 8r93, 8r94, 8r95, 8r9c, 8r9d, 8r9e, 8r9f, 8r9g, 8r9h, 8r9i, 8r9j, 8r9k, 8r9l, 8r9m, 8r9n, 8r9p, 8r9q, 8ra1, 8ra2, 8ra3, 8ra4, 8ra5, 8ra6, 8ra7, 8ra8, 8ra9, 8raa, 8rab, 8rac, 8rad and 8rae. CXIDB ID 213.
- Gihan Ketawala, Caitlin M. Reiter, Petra Fromme and Sabine Botha. "The Pixel Anomaly Detection Tool: a user-friendly GUI for classifying detector frames using machine-learning approaches". Journal of Applied Crystallography 57 (2024) 529-538. doi:10.1107/s1600576724000116 (open access - download PDF).
- Marina Galchenkova, Alexandra Tolstikova, Bjarne Klopprogge, Janina Sprenger, et al. "Data reduction in protein serial crystallography". IUCrJ 11 (2024) 190-201. doi:10.1107/s205225252400054x (open access - download PDF).
- Hongjie Li, Yoshiki Nakajima, Eriko Nango, Shigeki Owada, et al. "Oxygen-evolving photosystem II structures during S1–S2–S3 transitions". Nature 626 (2024) 670-677. doi:10.1038/s41586-023-06987-5 (open access - download PDF).
- Robert W. Henning, Irina Kosheleva, Vukica Šrajer, In-Sik Kim, et al. "BioCARS: Synchrotron facility for probing structural dynamics of biological macromolecules". Structural Dynamics 11 (2024). doi:10.1063/4.0000238
- Anaïs Chretien, Marius F. Nagel, Sabine Botha, Raphaël de Wijn, et al. "Light-induced Trpin/Metout Switching During BLUF Domain Activation in ATP-bound Photoactivatable Adenylate Cyclase OaPAC". Journal of Molecular Biology 436 (2024) 168439. doi:10.1016/j.jmb.2024.168439 (open access). PDB IDs 8qfi, 8qfh and 8qfj.
- Toshiki Higashino, Satoru Inoue, Shunto Arai, Seiji Tsuzuki, et al. "Effects of Thiophene-Fused Isomer on High-Layered Crystallinity in π-Extended and Alkylated Organic Semiconductors". Chemistry of Materials 36 (2024) 848-859. doi:10.1021/acs.chemmater.3c02500
2023 (38 articles and 117 PDB entries this year)
- Marcin Sikorski, Marco Ramilli, Raphael de Wijn, Viktoria Hinger, et al. "First operation of the JUNGFRAU detector in 16-memory cell mode at European XFEL". Frontiers in Physics 11 (2023). doi:10.3389/fphy.2023.1303247 (open access).
- Maximilian Wranik, Michal W. Kepa, Emma V. Beale, Daniel James, et al. "A multi-reservoir extruder for time-resolved serial protein crystallography and compound screening at X-ray free-electron lasers". Nature Communications 14 (2023). doi:10.1038/s41467-023-43523-5 (open access - download PDF). PDB IDs 8cld, 8cl7, 8cl9, 8cle, 8clg, 8cl8, 8clh, 8cl6, 8clb, 8clf, 8clc and 8cl5.
- Cecilia Safari, Swagatha Ghosh, Rebecka Andersson, Jonatan Johannesson, et al. "Time-resolved serial crystallography to track the dynamics of carbon monoxide in the active site of cytochrome c oxidase". Science Advances 9 (2023). doi:10.1126/sciadv.adh4179 PDB IDs 8ajz, 8k65 and 8k6y.
- Lainey J. Williamson, Marina Galchenkova, Hannah L. Best, Richard J. Bean, et al. "Structure of the Lysinibacillus sphaericus Tpp49Aa1 pesticidal protein elucidated from natural crystals using MHz-SFX". Proceedings of the National Academy of Sciences 120 (2023). doi:10.1073/pnas.2203241120 (open access - download PDF). PDB IDs 7qa1, 8bey, 8bex and 8bez.
- Pascal Hogan-Lamarre, Yi Luo, Robert Bücker, R. J. Dwayne Miller, et al. "STEM SerialED: achieving high-resolution data for ab initio structure determination of beam-sensitive nanocrystalline materials". IUCrJ 11 (2024) 62-72. doi:10.1107/s2052252523009661 (open access - download PDF).
- Nina-Eleni Christou, Virginia Apostolopoulou, Diogo V. M. Melo, Matthias Ruppert, et al. "Time-resolved crystallography captures light-driven DNA repair". Science 382 (2023) 1015-1020. doi:10.1126/science.adj4270 PDB IDs 8oet, 8oyb, 8oy9, 8oyc, 8oya, 8oy4, 8oy5, 8oy6, 8oy7, 8oy8 and 8oy3.
- Manuel Maestre-Reyna, Po-Hsun Wang, Eriko Nango, Yuhei Hosokawa, et al. "Visualizing the DNA repair process by a photolyase at atomic resolution". Science 382 (2023). doi:10.1126/science.add7795 PDB IDs 7yeb, 7ycm, 7yei, 7yd7, 7yd6, 7yee, 7yej, 7ycp, 7ye0, 7ydz, 7yel, 7yem, 7yec, 7ycr, 7yek, 7yd8 and 7yc7.
- Jihan Kim, Youngchang Kim, Jaehyun Park, Ki Hyun Nam, et al. "Structural mechanism of Escherichia coli cyanase". Acta Crystallographica Section D Structural Biology 79 (2023) 1094-1108. doi:10.1107/s2059798323009609 PDB ID 8k6u.
- Oleksii Turkot, Fabio Dall'Antonia, Richard J. Bean, Juncheng E, et al. "Towards automated analysis of serial crystallography data at European XFEL". X-Ray Free-Electron Lasers: Advances in Source Development and Instrumentation VI (2023) 45. doi:10.1117/12.2669569
- Junko Tanaka, Satoshi Abe, Tohru Hayakawa, Mariko Kojima, et al. "Crystal structure of the in-cell Cry1Aa purified from Bacillus thuringiensis". Biochemical and Biophysical Research Communications 685 (2023) 149144. doi:10.1016/j.bbrc.2023.149144
- Sergi Plana-Ruiz, Alejandro Gómez-Pérez, Monika Budayova-Spano, Daniel L. Foley, et al. "High-Resolution Electron Diffraction of Hydrated Protein Crystals at Room Temperature". ACS Nano 17 (2023) 24802-24813. doi:10.1021/acsnano.3c05378 (open access - download PDF).
- Yongsam Kim and Ki Hyun Nam. "Fixed-Target Pink-Beam Serial Synchrotron Crystallography at Pohang Light Source II". Crystals 13 (2023) 1544. doi:10.3390/cryst13111544 (open access - download PDF). PDB IDs 8wdi and 8wdh. Data deposition paper.
- Monika Bjelčić, Kajsa G. V. Sigfridsson Clauss, Oskar Aurelius, Mirko Milas, et al. "Anaerobic fixed-target serial crystallography using sandwiched silicon nitride membranes". Acta Crystallographica Section D Structural Biology 79 (2023) 1018-1025. doi:10.1107/s205979832300880x (open access - download PDF). PDB IDs 8puq and 8pur.
- Alisia Fadini, Christopher D.M. Hutchison, Dmitry Morozov, Jeffrey Chang, et al. "Serial Femtosecond Crystallography Reveals that Photoactivation in a Fluorescent Protein Proceeds via the Hula Twist Mechanism". Journal of the American Chemical Society 145 (2023) 15796-15808. doi:10.1021/jacs.3c02313 (open access - download PDF). PDB IDs 8a6q, 8a6n, 8a6g, 8a7v, 8a6r, 8a6o, 8a6s and 8a6p.
- Diandra Doppler, Mukul Sonker, Ana Egatz-Gomez, Alice Grieco, et al. "Modular droplet injector for sample conservation providing new structural insight for the conformational heterogeneity in the disease-associated NQO1 enzyme". Lab on a Chip 23 (2023) 3016-3033. doi:10.1039/d3lc00176h PDB IDs 8c9j and 8fwa.
- Thomas Gruhl, Tobias Weinert, Matthew J. Rodrigues, Christopher J. Milne, et al. "Ultrafast structural changes direct the first molecular events of vision". Nature 615 (2023) 939-944. doi:10.1038/s41586-023-05863-6 (open access - download PDF). PDB IDs 8a6e, 8a6d, 7zbc, 7zbe and 8a6c.
- Tek Narsingh Malla, Kara Zielinski, Luis Aldama, Sasa Bajt, et al. "Heterogeneity in M. tuberculosis β-lactamase inhibition by Sulbactam". Nature Communications 14 (2023). doi:10.1038/s41467-023-41246-1 (open access - download PDF). PDB IDs 8gcs, 8gcx, 8gcv, 8ec4, 8ebi, 8gct and 8ebr.
- Izumi Ishigami, Sergio Carbajo, Nadia Zatsepin, Masahide Hikita, et al. "Detection of a Geminate Photoproduct of Bovine Cytochrome c Oxidase by Time-Resolved Serial Femtosecond Crystallography". Journal of the American Chemical Society 145 (2023) 22305-22309. doi:10.1021/jacs.3c07803 PDB ID 8gbt.
- In Jung Kim, Soo Rin Kim, Kyoung Heon Kim, Uwe T. Bornscheuer, et al. "Characterization and structural analysis of the endo-1,4-β-xylanase GH11 from the hemicellulose-degrading Thermoanaerobacterium saccharolyticum useful for lignocellulose saccharification". Scientific Reports 13 (2023). doi:10.1038/s41598-023-44495-8 (open access - download PDF). PDB ID 8ih1. Data deposition paper.
- Filip Leonarski, Jie Nan, Zdenek Matej, Quentin Bertrand, et al. "Kilohertz serial crystallography with the JUNGFRAU detector at a fourth-generation synchrotron source". IUCrJ 10 (2023) 729-737. doi:10.1107/s2052252523008618 (open access - download PDF). PDB IDs 8p1a, 8p1b, 8p1c and 8p1d.
- Melissa Carrillo, Thomas J. Mason, Agnieszka Karpik, Isabelle Martiel, et al. "Micro-structured polymer fixed targets for serial crystallography at synchrotrons and XFELs". IUCrJ 10 (2023) 678-693. doi:10.1107/s2052252523007595 (open access - download PDF). PDB IDs 9rvi, 9rvo and 9rv6.
- Pedram Mehrabi, Sihyun Sung, David von Stetten, Andreas Prester, et al. "Millisecond cryo-trapping by the spitrobot crystal plunger simplifies time-resolved crystallography". Nature Communications 14 (2023). doi:10.1038/s41467-023-37834-w (open access - download PDF). PDB IDs 8awy and 8b3m.
- Alessandra Henkel, Marina Galchenkova, Julia Maracke, Oleksandr Yefanov, et al. "JINXED: just in time crystallization for easy structure determination of biological macromolecules". IUCrJ 10 (2023) 253-260. doi:10.1107/s2052252523001653 (open access - download PDF). PDB IDs 8b3l, 8b3v, 8b3t and 8b3u.
- Alexander M. Wolff, Eriko Nango, Iris D. Young, Aaron S. Brewster, et al. "Mapping protein dynamics at high spatial resolution with temperature-jump X-ray crystallography". Nature Chemistry 15 (2023) 1549-1558. doi:10.1038/s41557-023-01329-4 (open access - download PDF).
- Izumi Ishigami, Raymond G. Sierra, Zhen Su, Ariana Peck, et al. "Structural insights into functional properties of the oxidized form of cytochrome c oxidase". Nature Communications 14 (2023). doi:10.1038/s41467-023-41533-x (open access - download PDF). PDB ID 8gcq.
- Hoyoung Kim, Taehyun Lim, Go Eun Ha, Jee-Young Lee, et al. "Structure-based drug discovery of a corticotropin-releasing hormone receptor 1 antagonist using an X-ray free-electron laser". Experimental & Molecular Medicine 55 (2023) 2039-2050. doi:10.1038/s12276-023-01082-1 (open access - download PDF). PDB IDs 8gtg, 8gtm and 8gti.
- Christopher D. M. Hutchison, James M. Baxter, Ann Fitzpatrick, Gabriel Dorlhiac, et al. "Optical control of ultrafast structural dynamics in a fluorescent protein". Nature Chemistry 15 (2023) 1607-1615. doi:10.1038/s41557-023-01275-1 (open access - download PDF). PDB IDs 7qln, 7qlm and 7qlo.
- Marina Galchenkova, Aida Rahmani Mashhour, Patrick Y. A. Reinke, Sebastian Günther, et al. "An Optimized Approach for Serial Crystallography Using Chips". Crystals 13 (2023) 1225. doi:10.3390/cryst13081225 (open access - download PDF).
- Kirill Kovalev, Fedor Tsybrov, Alexey Alekseev, Vitaly Shevchenko, et al. "Mechanisms of inward transmembrane proton translocation". Nature Structural & Molecular Biology (2023). doi:10.1038/s41594-023-01020-9 PDB IDs 7zne, 7zng, 7znh and 7zni.
- Cecilia M. Casadei, Ahmad Hosseinizadeh, Spencer Bliven, Tobias Weinert, et al. "Low-pass spectral analysis of time-resolved serial femtosecond crystallography data". Structural Dynamics 10 (2023). doi:10.1063/4.0000178
- Ki Hyun Nam. "Real-time monitoring of large-scale crystal growth using batch crystallization for serial crystallography". Journal of Crystal Growth 614 (2023) 127219. doi:10.1016/j.jcrysgro.2023.127219
- Kunio Nakata, Tatsuki Kashiwagi, Naoki Kunishima, Hisashi Naitow, et al. "Ambient temperature structure of phosphoketolase from Bifidobacterium longum determined by serial femtosecond X-ray crystallography". Acta Crystallographica Section D Structural Biology 79 (2023) 290-303. doi:10.1107/s2059798323001638 PDB IDs 7c8h and 7c8i.
- Kiyofumi Takaba, Saori Maki-Yonekura, Ichiro Inoue, Kensuke Tono, et al. "Structural resolution of a small organic molecule by serial X-ray free-electron laser and electron crystallography". Nature Chemistry 15 (2023) 491-497. doi:10.1038/s41557-023-01162-9 (open access - download PDF). CXIDB ID 206. CCDC ID 2119567.
- Swagatha Ghosh, Doris Zorić, Peter Dahl, Monika Bjelčić, et al. "A simple goniometer-compatible flow cell for serial synchrotron X-ray crystallography". Journal of Applied Crystallography 56 (2023) 449-460. doi:10.1107/s1600576723001036 (open access - download PDF). PDB ID 8hua.
- Matthew J. Rodrigues, Cecilia M. Casadei, Tobias Weinert, Valerie Panneels, et al. "Correction of rhodopsin serial crystallography diffraction intensities for a lattice-translocation defect". Acta Crystallographica Section D Structural Biology 79 (2023) 224-233. doi:10.1107/s2059798323000931 (open access - download PDF).
- Maximilian Wranik, Tobias Weinert, Chavdar Slavov, Tiziana Masini, et al. "Watching the release of a photopharmacological drug from tubulin using time-resolved serial crystallography". Nature Communications 14 (2023). doi:10.1038/s41467-023-36481-5 (open access - download PDF). PDB IDs 8ql2, 7yz3, 7yz6, 7yz5, 7yyz, 7yyy, 8qlb, 8ql9, 7yz1, 7yz0, 7yz2 and 8qla.
- Feng-Zhu Zhao, Zhi-Jun Wang, Qing-Jie Xiao, Li Yu, et al. "Microfluidic rotating-target device capable of three-degrees-of-freedom motion for efficient in situ serial synchrotron crystallography". Journal of Synchrotron Radiation 30 (2023) 347-358. doi:10.1107/s1600577523000462 (open access - download PDF). PDB ID 7dln.
- Rebecca J. Jernigan, Dhenugen Logeswaran, Diandra Doppler, Nirupa Nagaratnam, et al. "Room-temperature structural studies of SARS-CoV-2 protein NendoU with an X-ray free-electron laser". Structure 31 (2023) 138-151.e5. doi:10.1016/j.str.2022.12.009 PDB ID 7k9p.
2022 (31 articles and 106 PDB entries this year)
- T. Barends, K. Nass, A. Gorel and I. Schlichting. "Lysozyme, 9-11 fs FEL pulses as determined by XTCAV". Worldwide Protein Data Bank (2023). doi:10.2210/pdb8a9e/pdb (no associated journal article). PDB ID 8a9e.
- T. Barends, K. Nass, A. Gorel and I. Schlichting. "Thaumatin, 9-11 fs FEL pulses as determined by XTCAV". Worldwide Protein Data Bank (2022). doi:10.2210/pdb8a9f/pdb (no associated journal article). PDB ID 8a9f.
- Esra Ayan, Busra Yuksel, Ebru Destan, Fatma Betul Ertem, et al. "Cooperative allostery and structural dynamics of streptavidin at cryogenic- and ambient-temperature". Communications Biology 5 (2022). doi:10.1038/s42003-021-02903-7 (open access - download PDF). PDB ID 7ek8.
- Scisung Chung, Mi-Sun Kang, Dauren S. Alimbetov, Gil-Im Mun, et al. "Regulation of BRCA1 stability through the tandem UBX domains of isoleucyl-tRNA synthetase 1". Nature Communications 13 (2022). doi:10.1038/s41467-022-34612-y (open access - download PDF). PDB ID 7wrs.
- Hasan DeMirci, Yashas Rao, Gabriele M. Stoffel, Bastian Vögeli, et al. "Intersubunit Coupling Enables Fast CO2-Fixation by Reductive Carboxylases". ACS Central Science 8 (2022) 1091-1101. doi:10.1021/acscentsci.2c00057 (open access - download PDF). PDB ID 6na6.
- Andrea Cellini, Madan Kumar Shankar, Weixiao Yuan Wahlgren, Amke Nimmrich, et al. "Structural basis of the radical pair state in photolyases and cryptochromes". Chemical Communications 58 (2022) 4889-4892. doi:10.1039/d2cc00376g (open access - download PDF). PDB ID 7qut.
- James M. Baxter, Christopher D. M. Hutchison, Karim Maghlaoui, Violeta Cordon-Preciado, et al. "Observation of Cation Chromophore Photoisomerization of a Fluorescent Protein Using Millisecond Synchrotron Serial Crystallography and Infrared Vibrational and Visible Spectroscopy". The Journal of Physical Chemistry B 126 (2022) 9288-9296. doi:10.1021/acs.jpcb.2c06780 (open access - download PDF). PDB IDs 7tsv, 7tsu, 7tsr and 7tss.
- Elizaveta Lyapina, Egor Marin, Anastasiia Gusach, Philipp Orekhov, et al. "Structural basis for receptor selectivity and inverse agonism in S1P5 receptors". Nature Communications 13 (2022). doi:10.1038/s41467-022-32447-1 (open access - download PDF). PDB ID 7yxa.
- Satoshi Abe, Junko Tanaka, Mariko Kojima, Shuji Kanamaru, et al. "Cell-free protein crystallization for nanocrystal structure determination". Scientific Reports 12 (2022). doi:10.1038/s41598-022-19681-9 (open access - download PDF). PDB IDs 7xhr and 7xws.
- Virgile Adam, Kyprianos Hadjidemetriou, Nickels Jensen, Robert L. Shoeman, et al. "Rational Control of Off‐State Heterogeneity in a Photoswitchable Fluorescent Protein Provides Switching Contrast Enhancement**". ChemPhysChem 23 (2022). doi:10.1002/cphc.202200192 (open access - download PDF). PDB IDs 7o7x, 7o7w, 7o7v and 7o7u.
- Takeshi Murakawa, Mamoru Suzuki, Kenji Fukui, Tetsuya Masuda, et al. "Serial femtosecond X-ray crystallography of an anaerobically formed catalytic intermediate of copper amine oxidase". Acta Crystallographica Section D Structural Biology 78 (2022) 1428-1438. doi:10.1107/s2059798322010385 PDB ID 7ynh.
- Alireza Sadri, Marjan Hadian-Jazi, Oleksandr Yefanov, Marina Galchenkova, et al. "Automatic bad-pixel mask maker for X-ray pixel detectors with application to serial crystallography". Journal of Applied Crystallography 55 (2022) 1549-1561. doi:10.1107/s1600576722009815 (open access - download PDF).
- Yongsam Kim and Ki Hyun Nam. "Pink-Beam Serial Synchrotron Crystallography at Pohang Light Source II". Crystals 12 (2022) 1637. doi:10.3390/cryst12111637 (open access - download PDF). PDB IDs 8h8t, 8h8u, 8h8v and 8h8w. Data deposition paper.
- Kara A Zielinski, Andreas Prester, Hina Andaleeb, Soi Bui, et al. "Rapid and efficient room-temperature serial synchrotron crystallography using the CFEL TapeDrive". IUCrJ 9 (2022) 778-791. doi:10.1107/s2052252522010193 (open access - download PDF). PDB IDs 8af7, 8af8, 8af4, 8af5, 8af6, 7zq0, 7qar and 7zpv.
- Marina Lučić, Michael T. Wilson, Takehiko Tosha, Hiroshi Sugimoto, et al. "Serial Femtosecond Crystallography Reveals the Role of Water in the One- or Two-Electron Redox Chemistry of Compound I in the Catalytic Cycle of the B-Type Dye-Decolorizing Peroxidase DtpB". ACS Catalysis 12 (2022) 13349-13359. doi:10.1021/acscatal.2c03754 (open access - download PDF). PDB IDs 7qzf, 7qzh, 7qze, 7qzg and 7zmj.
- Mukul Sonker, Diandra Doppler, Ana Egatz-Gomez, Sahba Zaare, et al. "Electrically stimulated droplet injector for reduced sample consumption in serial crystallography". Biophysical Reports 2 (2022) 100081. doi:10.1016/j.bpr.2022.100081 (open access).
- Kyprianos Hadjidemetriou, Nicolas Coquelle, Thomas R. M. Barends, Elke De Zitter, et al. "Time-resolved serial femtosecond crystallography on fatty-acid photodecarboxylase: lessons learned". Acta Crystallographica Section D Structural Biology 78 (2022) 1131-1142. doi:10.1107/s2059798322007525 (open access - download PDF). PDB IDs 7r33, 7r34, 7r35 and 7r36.
- Susannah Holmes, Henry J. Kirkwood, Richard Bean, Klaus Giewekemeyer, et al. "Megahertz pulse trains enable multi-hit serial femtosecond crystallography experiments at X-ray free electron lasers". Nature Communications 13 (2022). doi:10.1038/s41467-022-32434-6 (open access - download PDF). PDB IDs 6web, 7tum and 6wec.
- Guillaume Tetreau, Michael R. Sawaya, Elke De Zitter, Elena A. Andreeva, et al. "De novo determination of mosquitocidal Cry11Aa and Cry11Ba structures from naturally-occurring nanocrystals". Nature Communications 13 (2022). doi:10.1038/s41467-022-31746-x (open access - download PDF). PDB IDs 7qx4, 7qx5, 7qx6 and 7qx7. CXIDB ID 190.
- Keondo Lee, Jihan Kim, Sangwon Baek, Jaehyun Park, et al. "Combination of an inject-and-transfer system for serial femtosecond crystallography". Journal of Applied Crystallography 55 (2022) 813-822. doi:10.1107/s1600576722005556 PDB IDs 7wkr and 7wuc.
- Tadeo Moreno-Chicano, Leiah M. Carey, Danny Axford, John H. Beale, et al. "Complementarity of neutron, XFEL and synchrotron crystallography for defining the structures of metalloenzymes at room temperature". IUCrJ 9 (2022) 610-624. doi:10.1107/s2052252522006418 (open access - download PDF). PDB IDs 7adf and 7adx.
- Shangji Zhang, Debra T. Hansen, Jose M. Martin-Garcia, James D. Zook, et al. "Purification, characterization, and preliminary serial crystallography diffraction advances structure determination of full-length human particulate guanylyl cyclase A receptor". Scientific Reports 12 (2022). doi:10.1038/s41598-022-15798-z (open access - download PDF).
- Petra Båth, Analia Banacore, Per Börjesson, Robert Bosman, et al. "Lipidic cubic phase serial femtosecond crystallography structure of a photosynthetic reaction centre". Acta Crystallographica Section D Structural Biology 78 (2022) 698-708. doi:10.1107/s2059798322004144 (open access - download PDF). PDB IDs 7q7p and 7q7q.
- Ki Hyun Nam, Sehan Park and Jaehyun Park. "Preliminary XFEL data from spontaneously grown endo-1,4-β-xylanase crystals from Hypocrea virens". Acta Crystallographica Section F Structural Biology Communications 78 (2022) 226-231. doi:10.1107/s2053230x22005118
- Henry J. Kirkwood, Raphael de Wijn, Grant Mills, Romain Letrun, et al. "A multi-million image Serial Femtosecond Crystallography dataset collected at the European XFEL". Scientific Data 9 (2022). doi:10.1038/s41597-022-01266-w (open access - download PDF). CXIDB ID 185.
- Manuel Maestre-Reyna, Cheng-Han Yang, Eriko Nango, Wei-Cheng Huang, et al. "Serial crystallography captures dynamic control of sequential electron and proton transfer events in a flavoenzyme". Nature Chemistry 14 (2022) 677-685. doi:10.1038/s41557-022-00922-3 PDB IDs 7vj8, 7viw, 7vix, 7viy, 7viz, 7vj0, 7vj1, 7vj2, 7vj3, 7vj4, 7vj5, 7vj6, 7vj9, 7vja, 7vjb, 7vjc, 7vje, 7vjg, 7vjh, 7vji, 7vjj, 7vjk and 7vj7.
- Jose M. Martin-Garcia, Sabine Botha, Hao Hu, Rebecca Jernigan, et al. "Serial macromolecular crystallography at ALBA Synchrotron Light Source". Journal of Synchrotron Radiation 29 (2022) 896-907. doi:10.1107/s1600577522002508 (open access - download PDF). PDB IDs 7s50, 7s4w, 7s4y, 7s4z and 7s4r.
- Michal. W. Kepa, Takashi Tomizaki, Yohei Sato, Dmitry Ozerov, et al. "Acoustic levitation and rotation of thin films and their application for room temperature protein crystallography". Scientific Reports 12 (2022). doi:10.1038/s41598-022-09167-z (open access - download PDF).
- Toshiaki Hosaka, Takashi Nomura, Minoru Kubo, Takanori Nakane, et al. "Conformational alterations in unidirectional ion transport of a light-driven chloride pump revealed using X-ray free electron lasers". Proceedings of the National Academy of Sciences 119 (2022). doi:10.1073/pnas.2117433119 (open access - download PDF). PDB IDs 7vgt, 7vgu and 7vgv. CXIDB ID 192.
- Mohammad Vakili, Johan Bielecki, Juraj Knoška, Florian Otte, et al. "3D printed devices and infrastructure for liquid sample delivery at the European XFEL". Journal of Synchrotron Radiation 29 (2022) 331-346. doi:10.1107/s1600577521013370 (open access - download PDF).
- Sandra Mous, Guillaume Gotthard, David Ehrenberg, Saumik Sen, et al. "Dynamics and mechanism of a light-driven chloride pump". Science 375 (2022) 845-851. doi:10.1126/science.abj6663 PDB IDs 7o8f, 7o8g, 7o8h, 7o8i, 7o8j, 7o8k, 7o8l, 7o8m, 7o8n, 7o8o, 7o8p, 7o8q, 7o8r, 7o8s, 7o8t, 7o8u, 7o8v, 7o8y and 7o8z.
- Ki Hyun Nam. "Processing of Multicrystal Diffraction Patterns in Macromolecular Crystallography Using Serial Crystallography Programs". Crystals 12 (2022) 103. doi:10.3390/cryst12010103 (open access - download PDF). PDB IDs 7wbe and 7wbd.
- Ki Hyun Nam. "Beef tallow injection matrix for serial crystallography". Scientific Reports 12 (2022). doi:10.1038/s41598-021-04714-6 (open access - download PDF). PDB IDs 7e02 and 7e03. CXIDB IDs 165 and 166.
2021 (39 articles and 96 PDB entries this year)
- Y. Hyun and N.-C. Ha. "Crystal structure of Brucella abortus PhiA". Worldwide Protein Data Bank (2022). doi:10.2210/pdb7dsg/pdb (no associated journal article). PDB ID 7dsg.
- Halilibrahim Ciftci, Hiroshi Tateishi, Kotaro Koiwai, Ryoko Koga, et al. "Structural insight into host plasma membrane association and assembly of HIV-1 matrix protein". Scientific Reports 11 (2021). doi:10.1038/s41598-021-95236-8 (open access - download PDF). PDB ID 7e1i.
- Hanna Kwon, Jaswir Basran, Chinar Pathak, Mahdi Hussain, et al. "XFEL Crystal Structures of Peroxidase Compound II". Angewandte Chemie International Edition 60 (2021) 14578-14585. doi:10.1002/anie.202103010 (open access - download PDF). PDB IDs 7biu and 7bi1.
- Takashi Nomura, Tetsunari Kimura, Yusuke Kanematsu, Daichi Yamada, et al. "Short-lived intermediate in N 2 O generation by P450 NO reductase captured by time-resolved IR spectroscopy and XFEL crystallography". Proceedings of the National Academy of Sciences 118 (2021). doi:10.1073/pnas.2101481118 PDB ID 7dvo.
- P. Mehrabi, R. Bücker, G. Bourenkov, H.M. Ginn, et al. "Serial femtosecond and serial synchrotron crystallography can yield data of equivalent quality: A systematic comparison". Science Advances 7 (2021). doi:10.1126/sciadv.abf1380 (open access - download PDF). PDB IDs 7a45, 7a43, 7a44 and 7a42.
- Shinya Hanashima, Takanori Nakane and Eiichi Mizohata. "Heavy Atom Detergent/Lipid Combined X-ray Crystallography for Elucidating the Structure-Function Relationships of Membrane Proteins". Membranes 11 (2021) 823. doi:10.3390/membranes11110823 (open access - download PDF). PDB ID 7vso.
- Max T. B. Clabbers, Susannah Holmes, Timothy W. Muusse, Parimala R. Vajjhala, et al. "MyD88 TIR domain higher-order assembly interactions revealed by microcrystal electron diffraction and serial femtosecond crystallography". Nature Communications 12 (2021). doi:10.1038/s41467-021-22590-6 (open access - download PDF). PDB ID 7ber.
- Marie Luise Grünbein, Alexander Gorel, Lutz Foucar, Sergio Carbajo, et al. "Effect of X-ray free-electron laser-induced shockwaves on haemoglobin microcrystals delivered in a liquid jet". Nature Communications 12 (2021). doi:10.1038/s41467-021-21819-8 (open access - download PDF). PDB IDs 7aeu, 7aev and 7aet.
- Robert D. Healey, Shibom Basu, Anne-Sophie Humm, Cedric Leyrat, et al. "An automated platform for structural analysis of membrane proteins through serial crystallography". Cell Reports Methods 1 (2021) 100102. doi:10.1016/j.crmeth.2021.100102 (open access). PDB ID 6yxd.
- Omur Guven, Mehmet Gul, Esra Ayan, J Austin Johnson, et al. "Case Study of High-Throughput Drug Screening and Remote Data Collection for SARS-CoV-2 Main Protease by Using Serial Femtosecond X-ray Crystallography". Crystals 11 (2021) 1579. doi:10.3390/cryst11121579 (open access - download PDF). PDB IDs 7vjz, 7vjy, 7vk1, 7vk2, 7vk0, 7vk5, 7vk6, 7vk3, 7vjx, 7vk8, 7vjw, 7vk4 and 7vk7.
- Melissa Carrillo, Suraj Pandey, Juan Sanchez, Moraima Noda, et al. "High-resolution crystal structures of transient intermediates in the phytochrome photocycle". Structure 29 (2021) 743-754.e4. doi:10.1016/j.str.2021.03.004 PDB IDs 7jr5 and 7jri.
- Dongqing Pan, Ryo Oyama, Tomomi Sato, Takanori Nakane, et al. "Crystal structure of CmABCB1 multi-drug exporter in lipidic mesophase revealed by LCP-SFX". IUCrJ 9 (2021) 134-145. doi:10.1107/s2052252521011611 (open access - download PDF). PDB ID 7fc9. CXIDB ID 193.
- Karol Nass, Camila Bacellar, Claudio Cirelli, Florian Dworkowski, et al. "Pink-beam serial femtosecond crystallography for accurate structure-factor determination at an X-ray free-electron laser". IUCrJ 8 (2021) 905-920. doi:10.1107/s2052252521008046 (open access - download PDF). PDB IDs 7o44, 7o5k, 7o51, 7o5j and 7o53. CXIDB ID 180.
- Takeshi Murakawa, Mamoru Suzuki, Toshi Arima, Michihiro Sugahara, et al. "Microcrystal preparation for serial femtosecond X-ray crystallography of bacterial copper amine oxidase". Acta Crystallographica Section F Structural Biology Communications 77 (2021) 356-363. doi:10.1107/s2053230x21008967 PDB ID 7f8k.
- Marjan Hadian-Jazi, Alireza Sadri, Anton Barty, Oleksandr Yefanov, et al. "Data reduction for serial crystallography using a robust peak finder". Journal of Applied Crystallography 54 (2021) 1360-1378. doi:10.1107/s1600576721007317 (open access - download PDF).
- Suraj Pandey, George Calvey, Andrea M. Katz, Tek Narsingh Malla, et al. "Observation of substrate diffusion and ligand binding in enzyme crystals using high-repetition-rate mix-and-inject serial crystallography". IUCrJ 8 (2021) 878-895. doi:10.1107/s2052252521008125 (open access - download PDF). PDB IDs 7k8l, 7k8e, 7k8f, 7k8h and 7k8k. CXIDB ID 170.
- Isabelle Martiel, John H. Beale, Agnieszka Karpik, Chia-Ying Huang, et al. "Versatile microporous polymer-based supports for serial macromolecular crystallography". Acta Crystallographica Section D Structural Biology 77 (2021) 1153-1167. doi:10.1107/s2059798321007324 (open access - download PDF). PDB ID 7ai8.
- Serdar Durdagi, Çağdaş Dağ, Berna Dogan, Merve Yigin, et al. "Near-physiological-temperature serial crystallography reveals conformations of SARS-CoV-2 main protease active site for improved drug repurposing". Structure 29 (2021) 1382-1396.e6. doi:10.1016/j.str.2021.07.007 (open access). PDB IDs 7cwb and 7cwc.
- Heng Liu, R. N. V. Krishna Deepak, Anna Shiriaeva, Cornelius Gati, et al. "Molecular basis for lipid recognition by the prostaglandin D2receptor CRTH2". Proceedings of the National Academy of Sciences 118 (2021). doi:10.1073/pnas.2102813118 PDB ID 7m8w.
- Andrea Cellini, Weixiao Yuan Wahlgren, Léocadie Henry, Suraj Pandey, et al. "The three-dimensional structure of Drosophila melanogaster (6–4) photolyase at room temperature". Acta Crystallographica Section D Structural Biology 77 (2021) 1001-1009. doi:10.1107/s2059798321005830 (open access - download PDF). PDB ID 7azt.
- Zhen Su, Joshua Cantlon, Lacey Douthit, Max Wiedorn, et al. "Serial crystallography using automated drop dispensing". Journal of Synchrotron Radiation 28 (2021) 1386-1392. doi:10.1107/s1600577521006160 (open access - download PDF).
- Ki Hyun Nam and Yunje Cho. "Stable sample delivery in a viscous medium via a polyimide-based single-channel microfluidic chip for serial crystallography". Journal of Applied Crystallography 54 (2021) 1081-1087. doi:10.1107/s1600576721005720 PDB ID 7dtf. CXIDB ID 164.
- Ki Hyun Nam, Jihan Kim and Yunje Cho. "Polyimide mesh-based sample holder with irregular crystal mounting holes for fixed-target serial crystallography". Scientific Reports 11 (2021). doi:10.1038/s41598-021-92687-x (open access - download PDF). PDB ID 7dtb. CXIDB ID 163.
- J. Mia Lahey-Rudolph, Robert Schönherr, Miriam Barthelmess, Pontus Fischer, et al. "Fixed-target serial femtosecond crystallography using in cellulo grown microcrystals". IUCrJ 8 (2021) 665-677. doi:10.1107/s2052252521005297 (open access - download PDF). PDB IDs 7asi and 7asx.
- Robert Bücker, Pascal Hogan-Lamarre and R. J. Dwayne Miller. "Serial Electron Diffraction Data Processing With diffractem and CrystFEL". Frontiers in Molecular Biosciences 8 (2021). doi:10.3389/fmolb.2021.624264 (open access).
- Qiang Zhou, Zeng-Qiang Gao, Zheng Dong, Yu-Meng Jiang, et al. "A reference-based multi-lattice indexing method integrating prior information correction and iterative refinement in protein crystallography". Acta Crystallographica Section A Foundations and Advances 77 (2021) 277-288. doi:10.1107/s2053273321003521
- Brenna Norton-Baker, Pedram Mehrabi, Juliane Boger, Robert Schönherr, et al. "A simple vapor-diffusion method enables protein crystallization inside the HARE serial crystallography chip". Acta Crystallographica Section D Structural Biology 77 (2021) 820-834. doi:10.1107/s2059798321003855 (open access - download PDF). PDB IDs 7nje, 7njf, 7njg, 7njh, 7nji, 7njj and 7nkf.
- D. Sorigué, K. Hadjidemetriou, S. Blangy, G. Gotthard, et al. "Mechanism and dynamics of fatty acid photodecarboxylase". Science 372 (2021). doi:10.1126/science.abd5687 PDB ID 6zh7. CXIDB ID 177.
- Ki Hyun Nam. "Room-Temperature Structure of Xylitol-Bound Glucose Isomerase by Serial Crystallography: Xylitol Binding in the M1 Site Induces Release of Metal Bound in the M2 Site". International Journal of Molecular Sciences 22 (2021) 3892. doi:10.3390/ijms22083892 (open access - download PDF). PDB IDs 7dfj and 7dfk.
- Kazumasa Oda, Takashi Nomura, Takanori Nakane, Keitaro Yamashita, et al. "Time-resolved serial femtosecond crystallography reveals early structural changes in channelrhodopsin". eLife 10 (2021). doi:10.7554/elife.62389 (open access - download PDF). PDB IDs 7c86, 7e6y, 7e6z, 7e70, 7e71 and 7e6x. CXIDB ID 150.
- Ji-Hye Yun, Xuanxuan Li, Jianing Yue, Jae-Hyun Park, et al. "Early-stage dynamics of chloride ion–pumping rhodopsin revealed by a femtosecond X-ray laser". Proceedings of the National Academy of Sciences 118 (2021). doi:10.1073/pnas.2020486118 (open access - download PDF). PDB IDs 7crj, 7cri, 7crk, 7crl, 7crs, 7crt, 7crx and 7cry.
- Saminathan Ramakrishnan, Jason R. Stagno, Chelsie E. Conrad, Jienyu Ding, et al. "Synchronous RNA conformational changes trigger ordered phase transitions in crystals". Nature Communications 12 (2021). doi:10.1038/s41467-021-21838-5 (open access - download PDF). PDB IDs 6vwt and 6vwv.
- Inhyuk Nam, Chang-Ki Min, Bonggi Oh, Gyujin Kim, et al. "High-brightness self-seeded X-ray free-electron laser covering the 3.5 keV to 14.6 keV range". Nature Photonics 15 (2021) 435-441. doi:10.1038/s41566-021-00777-z PDB IDs 7byo, 7d01, 7d04, 7byp, 7d02 and 7d05. CXIDB ID 172.
- Kazuya Hasegawa, Seiki Baba, Takashi Kawamura, Masaki Yamamoto, et al. "Evaluation of the data-collection strategy for room-temperature micro-crystallography studied by serial synchrotron rotation crystallography combined with the humid air and glue-coating method". Acta Crystallographica Section D Structural Biology 77 (2021) 300-312. doi:10.1107/s2059798321001686 (open access - download PDF). PDB IDs 7cdn, 7cdk, 7cdm, 7cdo, 7cdp, 7cdq, 7cdr, 7cds, 7cdt and 7cdu.
- Dieter K. Schneider, Wuxian Shi, Babak Andi, Jean Jakoncic, et al. "FMX – the Frontier Microfocusing Macromolecular Crystallography Beamline at the National Synchrotron Light Source II". Journal of Synchrotron Radiation 28 (2021) 650-665. doi:10.1107/s1600577520016173 (open access - download PDF).
- Claudia Stohrer, Sam Horrell, Susanne Meier, Marta Sans, et al. "Homogeneous batch micro-crystallization of proteins from ammonium sulfate". Acta Crystallographica Section D Structural Biology 77 (2021) 194-204. doi:10.1107/s2059798320015454 (open access - download PDF).
- Marjan Hadian-Jazi, Peter Berntsen, Hugh Marman, Brian Abbey, et al. "Analysis of Multi-Hit Crystals in Serial Synchrotron Crystallography Experiments Using High-Viscosity Injectors". Crystals 11 (2021) 49. doi:10.3390/cryst11010049 (open access - download PDF).
- Marie L. Grünbein, Lutz Foucar, Alexander Gorel, Mario Hilpert, et al. "Observation of shock-induced protein crystal damage during megahertz serial femtosecond crystallography". Physical Review Research 3 (2021). doi:10.1103/physrevresearch.3.013046 (open access).
- Samuel L. Rose, Svetlana V. Antonyuk, Daisuke Sasaki, Keitaro Yamashita, et al. "An unprecedented insight into the catalytic mechanism of copper nitrite reductase from atomic-resolution and damage-free structures". Science Advances 7 (2021). doi:10.1126/sciadv.abd8523 (open access - download PDF). PDB IDs 6zas, 6zau and 6zaw.
- Do-Heon Gu, Cheolsoo Eo, Dong Tak Jeong, Jeong-Sun Kim, et al. "Synchrotron serial crystallography at 11C beamline of Pohang Light Source-II". Korean Society for Structural Biology 8 (2020) 93-98. doi:10.34184/kssb.2020.8.4.93
2020 (37 articles and 90 PDB entries this year)
- O. Aurelius, J. John, I. Martiel, M. Marsh, et al. "Structure of Ribonucleotide reductase R2 from Escherichia coli collected by femtosecond serial crystallography on a COC membrane". Worldwide Protein Data Bank (2020). doi:10.2210/pdb7bet/pdb (no associated journal article). PDB ID 7bet.
- I. Martiel, M. Marsh, L. Vera, C.Y. Huang, et al. "Structure of thaumatin collected by femtosecond serial crystallography on a COC membrane". Worldwide Protein Data Bank (2021). doi:10.2210/pdb7at6/pdb (no associated journal article). PDB ID 7at6.
- M. Schmidt and T. Malla. "SARS CoV-2 MAIN PROTEASE 3CLpro, ROOM TEMPERATURE, DAMAGE FREE XFEL MONOCLINIC STRUCTURE". Worldwide Protein Data Bank (2020). doi:10.2210/pdb7jvz/pdb (no associated journal article). PDB ID 7jvz.
- Elin Claesson, Weixiao Yuan Wahlgren, Heikki Takala, Suraj Pandey, et al. "The primary structural photoresponse of phytochrome proteins captured by a femtosecond X-ray laser". eLife 9 (2020). doi:10.7554/elife.53514 (open access - download PDF). PDB IDs 6t3u and 6t3l.
- Dohyun Im, Asuka Inoue, Takaaki Fujiwara, Takanori Nakane, et al. "Structure of the dopamine D2 receptor in complex with the antipsychotic drug spiperone". Nature Communications 11 (2020). doi:10.1038/s41467-020-20221-0 (open access - download PDF). PDB ID 7dfp. CXIDB ID 110.
- Alexander Gorel, Marie Grünbein, Richard Bean, Johan Bielecki, et al. "Shock Damage Analysis in Serial Femtosecond Crystallography Data Collected at MHz X-ray Free-Electron Lasers". Crystals 10 (2020) 1145. doi:10.3390/cryst10121145 (open access - download PDF). CXIDB ID 144.
- Robert Dods, Petra Båth, Dmitry Morozov, Viktor Ahlberg Gagnér, et al. "Ultrafast structural changes within a photosynthetic reaction centre". Nature 589 (2020) 310-314. doi:10.1038/s41586-020-3000-7 PDB IDs 6zhw, 6zi4, 6zid, 6zi6, 6zi5, 6zi9 and 6zia.
- Stefan Görlich, Abisheik John Samuel, Richard Johannes Best, Ronald Seidel, et al. "Natural hybrid silica/protein superstructure at atomic resolution". Proceedings of the National Academy of Sciences 117 (2020) 31088-31093. doi:10.1073/pnas.2019140117 (open access - download PDF). PDB ID 6zq3.
- Egor Marin, Aleksandra Luginina, Anastasiia Gusach, Kirill Kovalev, et al. "Small-wedge synchrotron and serial XFEL datasets for Cysteinyl leukotriene GPCRs". Scientific Data 7 (2020). doi:10.1038/s41597-020-00729-2 (open access - download PDF). CXIDB IDs 106 and 107.
- Kentaro Ihara, Masakatsu Hato, Takanori Nakane, Keitaro Yamashita, et al. "Isoprenoid-chained lipid EROCOC17+4: a new matrix for membrane protein crystallization and a crystal delivery medium in serial femtosecond crystallography". Scientific Reports 10 (2020). doi:10.1038/s41598-020-76277-x (open access - download PDF). PDB IDs 6lpj and 6lpk. CXIDB ID 140.
- Isabelle Martiel, Chia-Ying Huang, Pablo Villanueva-Perez, Ezequiel Panepucci, et al. "Low-dose in situ prelocation of protein microcrystals by 2D X-ray phase-contrast imaging for serial crystallography". IUCrJ 7 (2020) 1131-1141. doi:10.1107/s2052252520013238 (open access - download PDF). PDB IDs 6yoc, 6yoe and 6yog.
- Feng-Zhu Zhao, Bo Sun, Li Yu, Qing-Jie Xiao, et al. "A novel sample delivery system based on circular motion for in situ serial synchrotron crystallography". Lab on a Chip 20 (2020) 3888-3898. doi:10.1039/d0lc00443j PDB IDs 7c09 and 7c0p.
- Ming-Yue Lee, James Geiger, Andrii Ishchenko, Gye Won Han, et al. "Harnessing the power of an X-ray laser for serial crystallography of membrane proteins crystallized in lipidic cubic phase". IUCrJ 7 (2020) 976-984. doi:10.1107/s2052252520012701 (open access - download PDF). PDB ID 6wqa.
- Suk-Youl Park, Hyeongju Choi, Cheolsoo Eo, Yunje Cho, et al. "Fixed-Target Serial Synchrotron Crystallography Using Nylon Mesh and Enclosed Film-Based Sample Holder". Crystals 10 (2020) 803. doi:10.3390/cryst10090803 (open access - download PDF). PDB IDs 7cvj, 7cvk, 7cvl and 7cvm. CXIDB IDs 157, 158, 159 and 160.
- Marina Lučić, Dimitri A. Svistunenko, Michael T. Wilson, Amanda K. Chaplin, et al. "Serial Femtosecond Zero Dose Crystallography Captures a Water‐Free Distal Heme Site in a Dye‐Decolorising Peroxidase to Reveal a Catalytic Role for an Arginine in FeIV=O Formation". Angewandte Chemie International Edition 59 (2020) 21656-21662. doi:10.1002/anie.202008622 (open access - download PDF). PDB IDs 6yrd and 6yrj.
- Austin Echelmeier, Jorvani Cruz Villarreal, Marc Messerschmidt, Daihyun Kim, et al. "Segmented flow generator for serial crystallography at the European X-ray free electron laser". Nature Communications 11 (2020). doi:10.1038/s41467-020-18156-7 (open access - download PDF). PDB ID 6u57. CXIDB ID 152.
- Karol Nass, Robert Cheng, Laura Vera, Aldo Mozzanica, et al. "Advances in long-wavelength native phasing at X-ray free-electron lasers". IUCrJ 7 (2020) 965-975. doi:10.1107/s2052252520011379 (open access - download PDF). PDB IDs 6s0l, 6s0q, 6s19, 6s1d, 6s1e and 6s1g. CXIDB IDs 103, 104 and 105.
- Anastasya Shilova, Hugo Lebrette, Oskar Aurelius, Jie Nan, et al. "Current status and future opportunities for serial crystallography at MAX IV Laboratory". Journal of Synchrotron Radiation 27 (2020) 1095-1102. doi:10.1107/s1600577520008735 (open access - download PDF). PDB IDs 6y4c, 6y78 and 6y2n.
- Ki Hyun Nam. "Lard Injection Matrix for Serial Crystallography". International Journal of Molecular Sciences 21 (2020) 5977. doi:10.3390/ijms21175977 (open access - download PDF). PDB IDs 7cjz and 7ck0. CXIDB IDs 153 and 154.
- Thomas Clairfeuille, Kerry R. Buchholz, Qingling Li, Erik Verschueren, et al. "Structure of the essential inner membrane lipopolysaccharide–PbgA complex". Nature 584 (2020) 479-483. doi:10.1038/s41586-020-2597-x
- Keondo Lee, Donghyeon Lee, Sangwon Baek, Jaehyun Park, et al. "Viscous-medium-based crystal support in a sample holder for fixed-target serial femtosecond crystallography". Journal of Applied Crystallography 53 (2020) 1051-1059. doi:10.1107/s1600576720008663 PDB IDs 6ll2 and 6ll3.
- Yingchen Shi and Haiguang Liu. "EM-detwin: A Program for Resolving Indexing Ambiguity in Serial Crystallography Using the Expectation-Maximization Algorithm". Crystals 10 (2020) 588. doi:10.3390/cryst10070588 (open access - download PDF).
- Nirupa Nagaratnam, Yanyang Tang, Sabine Botha, Justin Saul, et al. "Enhanced X-ray diffraction of in vivo-grown μNS crystals by viscous jets at XFELs". Acta Crystallographica Section F Structural Biology Communications 76 (2020) 278-289. doi:10.1107/s2053230x20006172 (open access - download PDF).
- Petr Skopintsev, David Ehrenberg, Tobias Weinert, Daniel James, et al. "Femtosecond-to-millisecond structural changes in a light-driven sodium pump". Nature 583 (2020) 314-318. doi:10.1038/s41586-020-2307-8 PDB IDs 6tk7, 6tk6, 6tk5, 6tk4, 6tk3, 6tk2 and 6tk1.
- Ki Hyun Nam. "Polysaccharide-Based Injection Matrix for Serial Crystallography". International Journal of Molecular Sciences 21 (2020) 3332. doi:10.3390/ijms21093332 (open access - download PDF). PDB IDs 7bvl, 7bvm, 7bvn and 7bvo. CXIDB IDs 124, 125, 126 and 127.
- Karol Nass, Alexander Gorel, Malik M. Abdullah, Andrew V. Martin, et al. "Structural dynamics in proteins induced by and probed with X-ray free-electron laser pulses". Nature Communications 11 (2020). doi:10.1038/s41467-020-15610-4 (open access - download PDF). PDB IDs 6srj, 6srq, 6srk, 6srl, 6sro, 6srp, 6sr0, 6sr1, 6sr2, 6sr3, 6sr4 and 6sr5.
- Xuanxuan Li, Chufeng Li and Haiguang Liu. "SPIND-TC: an indexing method for two-color X-ray diffraction data". Acta Crystallographica Section A Foundations and Advances 76 (2020) 369-375. doi:10.1107/s2053273320001916 (open access - download PDF).
- Donghyeon Lee, Sehan Park, Keondo Lee, Jangwoo Kim, et al. "Application of a high-throughput microcrystal delivery system to serial femtosecond crystallography". Journal of Applied Crystallography 53 (2020) 477-485. doi:10.1107/s1600576720002423 (open access - download PDF). PDB ID 6j43.
- James Zook, Mrinal Shekhar, Debra Hansen, Chelsie Conrad, et al. "XFEL and NMR Structures of Francisella Lipoprotein Reveal Conformational Space of Drug Target against Tularemia". Structure 28 (2020) 540-547.e3. doi:10.1016/j.str.2020.02.005 PDB ID 6pny.
- Alexander M. Wolff, Iris D. Young, Raymond G. Sierra, Aaron S. Brewster, et al. "Comparing serial X-ray crystallography and microcrystal electron diffraction (MicroED) as methods for routine structure determination from small macromolecular crystals". IUCrJ 7 (2020) 306-323. doi:10.1107/s205225252000072x (open access - download PDF). PDB ID 6u5e.
- Robert Bücker, Pascal Hogan-Lamarre, Pedram Mehrabi, Eike C. Schulz, et al. "Serial protein crystallography in an electron microscope". Nature Communications 11 (2020). doi:10.1038/s41467-020-14793-0 (open access - download PDF). PDB IDs 6s2o and 6s2n.
- Eugenio de la Mora, Nicolas Coquelle, Charles S. Bury, Martin Rosenthal, et al. "Radiation damage and dose limits in serial synchrotron crystallography at cryo- and room temperatures". Proceedings of the National Academy of Sciences 117 (2020) 4142-4151. doi:10.1073/pnas.1821522117 (open access - download PDF). PDB IDs 6q88 and 6q8t.
- Joyce Woodhouse, Gabriela Nass Kovacs, Nicolas Coquelle, Lucas M. Uriarte, et al. "Photoswitching mechanism of a fluorescent protein revealed by time-resolved crystallography and transient absorption spectroscopy". Nature Communications 11 (2020). doi:10.1038/s41467-020-14537-0 (open access - download PDF). PDB IDs 6t39 and 6t3a.
- Kirill Kovalev, Roman Astashkin, Ivan Gushchin, Philipp Orekhov, et al. "Molecular mechanism of light-driven sodium pumping". Nature Communications 11 (2020). doi:10.1038/s41467-020-16032-y (open access - download PDF). PDB ID 6yc0. CXIDB ID 141.
- Juraj Knoška, Luigi Adriano, Salah Awel, Kenneth R. Beyerlein, et al. "Ultracompact 3D microfluidics for time-resolved structural biology". Nature Communications 11 (2020). doi:10.1038/s41467-020-14434-6 (open access - download PDF). PDB ID 6r2o. CXIDB ID 120.
- Michihiro Sugahara, Koji Motomura, Mamoru Suzuki, Tetsuya Masuda, et al. "Viscosity-adjustable grease matrices for serial nanocrystallography". Scientific Reports 10 (2020). doi:10.1038/s41598-020-57675-7 (open access - download PDF). PDB IDs 6k2p, 6k2r, 6k2s, 6k2t, 6k2v, 6k2w and 6k2x. CXIDB ID 101.
- Karol Nass, Lars Redecke, M. Perbandt, O. Yefanov, et al. "In cellulo crystallization of Trypanosoma brucei IMP dehydrogenase enables the identification of genuine co-factors". Nature Communications 11 (2020). doi:10.1038/s41467-020-14484-w (open access - download PDF). PDB ID 6rfu.
- Diana C. F. Monteiro, David von Stetten, Claudia Stohrer, Marta Sans, et al. "3D-MiXD: 3D-printed X-ray-compatible microfluidic devices for rapid, low-consumption serial synchrotron crystallography data collection in flow". IUCrJ 7 (2020) 207-219. doi:10.1107/s2052252519016865 (open access - download PDF). PDB IDs 6rxh and 6rxi.
- Yaroslav Gevorkov, Anton Barty, Wolfgang Brehm, Thomas A. White, et al. "pinkIndexer – a universal indexer for pink-beam X-ray and electron diffraction snapshots". Acta Crystallographica Section A Foundations and Advances 76 (2020) 121-131. doi:10.1107/s2053273319015559 (open access - download PDF).
- Ki Hyun Nam. "Shortening injection matrix for serial crystallography". Scientific Reports 10 (2020). doi:10.1038/s41598-019-56135-1 (open access - download PDF). PDB IDs 6kca, 6kcb, 6kcc and 6kcd. CXIDB IDs 122, 113, 114 and 115.
2019 (39 articles and 98 PDB entries this year)
- Gabriela Nass Kovacs, Jacques-Philippe Colletier, Marie Luise Grünbein, Yang Yang, et al. "Three-dimensional view of ultrafast dynamics in photoexcited bacteriorhodopsin". Nature Communications 10 (2019). doi:10.1038/s41467-019-10758-0 (open access - download PDF). PDB IDs 6gai, 6gah, 6ga2, 6gag, 6gaf, 6gae, 6gad, 6gac, 6gab, 6gaa, 6ga9, 6ga8, 6ga7, 6ga6, 6ga5, 6ga4, 6ga3, 6ga1 and 6rmk.
- Izumi Ishigami, Ariel Lewis-Ballester, Austin Echelmeier, Gerrit Brehm, et al. "Snapshot of an oxygen intermediate in the catalytic reaction of cytochromecoxidase". Proceedings of the National Academy of Sciences 116 (2019) 3572-3577. doi:10.1073/pnas.1814526116 PDB IDs 6nmp, 6nmf and 6nkn.
- Tobias Weinert, Petr Skopintsev, Daniel James, Florian Dworkowski, et al. "Proton uptake mechanism in bacteriorhodopsin captured by serial synchrotron crystallography". Science 365 (2019) 61-65. doi:10.1126/science.aaw8634 PDB IDs 6rqp, 6rqo, 6rph and 6rnj.
- Ki Hyun Nam. "Stable sample delivery in viscous media via a capillary for serial crystallography". Journal of Applied Crystallography 53 (2020) 45-50. doi:10.1107/s1600576719014985 PDB IDs 6kd1 and 6kd2.
- M. L. Shelby, D. Gilbile, T. D. Grant, C. Seuring, et al. "A fixed-target platform for serial femtosecond crystallography in a hydrated environment". IUCrJ 7 (2020) 30-41. doi:10.1107/s2052252519014003 (open access - download PDF).
- Oleksandr Yefanov, Dominik Oberthür, Richard Bean, Max O. Wiedorn, et al. "Evaluation of serial crystallographic structure determination within megahertz pulse trains". Structural Dynamics 6 (2019). doi:10.1063/1.5124387 CXIDB ID 98.
- Suraj Pandey, Richard Bean, Tokushi Sato, Ishwor Poudyal, et al. "Time-resolved serial femtosecond crystallography at the European XFEL". Nature Methods 17 (2019) 73-78. doi:10.1038/s41592-019-0628-z PDB IDs 6p4i, 6p5d, 6p5e, 6p5g and 6p5f. CXIDB ID 100.
- Si Hoon Park, Jaehyun Park, Sang Jae Lee, Woo Seok Yang, et al. "A host dTMP-bound structure of T4 phage dCMP hydroxymethylase mutant using an X-ray free electron laser". Scientific Reports 9 (2019). doi:10.1038/s41598-019-52825-y (open access - download PDF). PDB ID 6l18.
- Chris Gisriel, Jesse Coe, Romain Letrun, Oleksandr M. Yefanov, et al. "Membrane protein megahertz crystallography at the European XFEL". Nature Communications 10 (2019). doi:10.1038/s41467-019-12955-3 (open access - download PDF). PDB ID 6pgk. CXIDB ID 111.
- Natasha Stander, Petra Fromme and Nadia Zatsepin. "DatView: a graphical user interface for visualizing and querying large data sets in serial femtosecond crystallography". Journal of Applied Crystallography 52 (2019) 1440-1448. doi:10.1107/s1600576719012044 (open access - download PDF).
- Andrii Ishchenko, Benjamin Stauch, Gye Won Han, Alexander Batyuk, et al. "Toward G protein-coupled receptor structure-based drug design using X-ray lasers". IUCrJ 6 (2019) 1106-1119. doi:10.1107/s2052252519013137 (open access - download PDF). PDB IDs 6prz, 6ps2, 6ps0, 6ps1, 6ps6, 6ps3, 6ps4, 6ps5, 6ps7 and 6ps8.
- Michihiro Suga, Fusamichi Akita, Keitaro Yamashita, Yoshiki Nakajima, et al. "An oxyl/oxo mechanism for oxygen-oxygen coupling in PSII revealed by an x-ray free-electron laser". Science 366 (2019) 334-338. doi:10.1126/science.aax6998 PDB IDs 6jlj, 6jlk, 6jll, 6jlm, 6jln, 6jlo and 6jlp.
- Yoshiaki Shimazu, Kensuke Tono, Tomoyuki Tanaka, Yasuaki Yamanaka, et al. "High-viscosity sample-injection device for serial femtosecond crystallography at atmospheric pressure". Journal of Applied Crystallography 52 (2019) 1280-1288. doi:10.1107/s1600576719012846 (open access - download PDF). PDB IDs 6jzh and 6jzi.
- Tadeo Moreno-Chicano, Ali Ebrahim, Danny Axford, Martin V. Appleby, et al. "High-throughput structures of protein–ligand complexes at room temperature using serial femtosecond crystallography". IUCrJ 6 (2019) 1074-1085. doi:10.1107/s2052252519011655 (open access - download PDF). PDB IDs 6i7f, 6i6g, 6i7c and 6qwg.
- Kotaro Koiwai, Jun Tsukimoto, Tetsuya Higashi, Fumitaka Mafuné, et al. "Improvement of Production and Isolation of Human Neuraminidase-1 in Cellulo Crystals". ACS Applied Bio Materials 2 (2019) 4941-4952. doi:10.1021/acsabm.9b00686
- Aleksandra Luginina, Anastasiia Gusach, Egor Marin, Alexey Mishin, et al. "Structure-based mechanism of cysteinyl leukotriene receptor inhibition by antiasthmatic drugs". Science Advances 5 (2019). doi:10.1126/sciadv.aax2518 PDB ID 6rz5.
- Rebecka Andersson, Cecilia Safari, Petra Båth, Robert Bosman, et al. "Well-based crystallization of lipidic cubic phase microcrystals for serial X-ray crystallography experiments". Acta Crystallographica Section D Structural Biology 75 (2019) 937-946. doi:10.1107/s2059798319012695 (open access - download PDF).
- Juan C. Sanchez, Melissa Carrillo, Suraj Pandey, Moraima Noda, et al. "High-resolution crystal structures of a myxobacterial phytochrome at cryo and room temperatures". Structural Dynamics 6 (2019). doi:10.1063/1.5120527 PDB ID 6ptq.
- Pedram Mehrabi, Eike C. Schulz, Michael Agthe, Sam Horrell, et al. "Liquid application method for time-resolved analyses by serial synchrotron crystallography". Nature Methods 16 (2019) 979-982. doi:10.1038/s41592-019-0553-1 PDB IDs 6rnf, 6rnd, 6rnc and 6rnb.
- Austin Echelmeier, Daihyun Kim, Jorvani Cruz Villarreal, Jesse Coe, et al. "3D printed droplet generation devices for serial femtosecond crystallography enabled by surface coating". Journal of Applied Crystallography 52 (2019) 997-1008. doi:10.1107/s1600576719010343
- Yaroslav Gevorkov, Oleksandr Yefanov, Anton Barty, Thomas A. White, et al. "XGANDALF – extended gradient descent algorithm for lattice finding". Acta Crystallographica Section A Foundations and Advances 75 (2019) 694-704. doi:10.1107/s2053273319010593 (open access - download PDF).
- P. Berntsen, M. Hadian Jazi, M. Kusel, A. V. Martin, et al. "The serial millisecond crystallography instrument at the Australian Synchrotron incorporating the “Lipidico” injector". Review of Scientific Instruments 90 (2019). doi:10.1063/1.5104298 PDB ID 6mqv.
- A. Tolstikova, M. Levantino, O. Yefanov, V. Hennicke, et al. "1 kHz fixed-target serial crystallography using a multilayer monochromator and an integrating pixel detector". IUCrJ 6 (2019) 927-937. doi:10.1107/s205225251900914x (open access - download PDF). PDB IDs 6qy2, 6qy1, 6qxw, 6qxv, 6qy4, 6qy0, 6qy5, 6qxy and 6qxx.
- Suk-Youl Park and Ki Hyun Nam. "Sample delivery using viscous media, a syringe and a syringe pump for serial crystallography". Journal of Synchrotron Radiation 26 (2019) 1815-1819. doi:10.1107/s160057751900897x PDB IDs 6jxp and 6jxq. CXIDB IDs 95 and 96.
- Robert J. Trachman, Jason R. Stagno, Chelsie Conrad, Christopher P. Jones, et al. "Co-crystal structure of the iMango-III fluorescent RNA aptamer using an X-ray free-electron laser". Acta Crystallographica Section F Structural Biology Communications 75 (2019) 547-551. doi:10.1107/s2053230x19010136 PDB ID 6pq7.
- P. Lindenberg, L. Ruiz Arana, L. K. Mahnke, P. Rönfeldt, et al. "New insights into the crystallization of polymorphic materials: from real-time serial crystallography to luminescence analysis". Reaction Chemistry & Engineering 4 (2019) 1757-1767. doi:10.1039/c9re00191c (open access - download PDF).
- Christopher Jones, Brandon Tran, Chelsie Conrad, Jason Stagno, et al. "Co-crystal structure of the Fusobacterium ulcerans ZTP riboswitch using an X-ray free-electron laser". Acta Crystallographica Section F Structural Biology Communications 75 (2019) 496-500. doi:10.1107/s2053230x19008549 PDB ID 6od9.
- Thomas P. Halsted, Keitaro Yamashita, Chai C. Gopalasingam, Rajesh T. Shenoy, et al. "Catalytically important damage-free structures of a copper nitrite reductase obtained by femtosecond X-ray laser and room-temperature neutron crystallography". IUCrJ 6 (2019) 761-772. doi:10.1107/s2052252519008285 (open access - download PDF). PDB IDs 6gsq, 6gt0 and 6gt2.
- Julia Lieske, Maximilian Cerv, Stefan Kreida, Dana Komadina, et al. "On-chip crystallization for serial crystallography experiments and on-chip ligand-binding studies". IUCrJ 6 (2019) 714-728. doi:10.1107/s2052252519007395 (open access - download PDF). PDB ID 6qf5.
- Soichiro Tsujino, Akira Shinoda and Takashi Tomizaki. "On-demand droplet loading of ultrasonic acoustic levitator and its application for protein crystallography experiments". Applied Physics Letters 114 (2019). doi:10.1063/1.5095574
- Donghyeon Lee, Sangwon Baek, Jaehyun Park, Keondo Lee, et al. "Nylon mesh-based sample holder for fixed-target serial femtosecond crystallography". Scientific Reports 9 (2019). doi:10.1038/s41598-019-43485-z (open access - download PDF). PDB IDs 6irj and 6irk. CXIDB IDs 89 and 90.
- Ali Ebrahim, Tadeo Moreno-Chicano, Martin V. Appleby, Amanda K. Chaplin, et al. "Dose-resolved serial synchrotron and XFEL structures of radiation-sensitive metalloproteins". IUCrJ 6 (2019) 543-551. doi:10.1107/s2052252519003956 (open access - download PDF). PDB ID 6i43.
- Linda C. Johansson, Benjamin Stauch, John D. McCorvy, Gye Won Han, et al. "XFEL structures of the human MT2 melatonin receptor reveal the basis of subtype selectivity". Nature 569 (2019) 289-292. doi:10.1038/s41586-019-1144-0 PDB IDs 6me6, 6me7, 6me8 and 6me9.
- Benjamin Stauch, Linda C. Johansson, John D. McCorvy, Nilkanth Patel, et al. "Structural basis of ligand recognition at the human MT1 melatonin receptor". Nature 569 (2019) 284-288. doi:10.1038/s41586-019-1141-3 PDB IDs 6me2, 6me3, 6me4 and 6me5.
- Jae-Hyun Park, Ji-Hye Yun, Yingchen Shi, Jeongmin Han, et al. "Non-Cryogenic Structure and Dynamics of HIV-1 Integrase Catalytic Core Domain by X-ray Free-Electron Lasers". International Journal of Molecular Sciences 20 (2019) 1943. doi:10.3390/ijms20081943 (open access - download PDF). PDB ID 6jcg. CXIDB ID 99.
- Jose M. Martin-Garcia, Lan Zhu, Derek Mendez, Ming-Yue Lee, et al. "High-viscosity injector-based pink-beam serial crystallography of microcrystals at a synchrotron radiation source". IUCrJ 6 (2019) 412-425. doi:10.1107/s205225251900263x (open access - download PDF). PDB ID 6mh8.
- Halil I Ciftci, Raymond G Sierra, Chun Hong Yoon, Zhen Su, et al. "Serial Femtosecond X-Ray Diffraction of HIV-1 Gag MA-IP6 Microcrystals at Ambient Temperature". International Journal of Molecular Sciences 20 (2019) 1675. doi:10.3390/ijms20071675 (open access - download PDF).
- Jaehyun Park, Sehan Park, Jangwoo Kim, Gisu Park, et al. "Polyacrylamide injection matrix for serial femtosecond crystallography". Scientific Reports 9 (2019). doi:10.1038/s41598-019-39020-9 (open access - download PDF). PDB IDs 6ig6 and 6ig7. CXIDB IDs 84 and 85.
- Diana C. F. Monteiro, Mohammad Vakili, Jessica Harich, Michael Sztucki, et al. "A microfluidic flow-focusing device for low sample consumption serial synchrotron crystallography experiments in liquid flow". Journal of Synchrotron Radiation 26 (2019) 406-412. doi:10.1107/s1600577519000304 PDB ID 6h79.
2018 (16 articles and 30 PDB entries this year)
- E. Han Dao, Frédéric Poitevin, Raymond G. Sierra, Cornelius Gati, et al. "Structure of the 30S ribosomal decoding complex at ambient temperature". RNA 24 (2018) 1667-1676. doi:10.1261/rna.067660.118 PDB ID 6cao.
- Chufeng Li, Xuanxuan Li, Richard Kirian, John C. H. Spence, et al. "SPIND: a reference-based auto-indexing algorithm for sparse serial crystallography data". IUCrJ 6 (2019) 72-84. doi:10.1107/s2052252518014951 (open access - download PDF).
- Martin Audet, Kate L. White, Billy Breton, Barbara Zarzycka, et al. "Crystal structure of misoprostol bound to the labor inducer prostaglandin E2 receptor". Nature Chemical Biology 15 (2018) 11-17. doi:10.1038/s41589-018-0160-y PDB ID 6m9t.
- Ji-Hye Yun, Xuanxuan Li, Jae-Hyun Park, Yang Wang, et al. "Non-cryogenic structure of a chloride pump provides crucial clues to temperature-dependent channel transport efficiency". Journal of Biological Chemistry 294 (2019) 794-804. doi:10.1074/jbc.ra118.004038 (open access). PDB ID 5ztl.
- R. Bruce Doak, Gabriela Nass Kovacs, Alexander Gorel, Lutz Foucar, et al. "Crystallography on a chip – without the chip: sheet-on-sheet sandwich". Acta Crystallographica Section D Structural Biology 74 (2018) 1000-1007. doi:10.1107/s2059798318011634 (open access - download PDF). PDB IDs 6gf0 and 6hal.
- Max O. Wiedorn, Dominik Oberthür, Richard Bean, Robin Schubert, et al. "Megahertz serial crystallography". Nature Communications 9 (2018). doi:10.1038/s41467-018-06156-7 (open access - download PDF). PDB IDs 6ftr and 6gth. CXIDB IDs 80 and 83.
- Nicole C. Woitowich, Andrei S. Halavaty, Patricia Waltz, Christopher Kupitz, et al. "Structural basis for light control of cell development revealed by crystal structures of a myxobacterial phytochrome". IUCrJ 5 (2018) 619-634. doi:10.1107/s2052252518010631 (open access - download PDF). PDB ID 6bay.
- Marie Luise Grünbein, Johan Bielecki, Alexander Gorel, Miriam Stricker, et al. "Megahertz data collection from protein microcrystals at an X-ray free-electron laser". Nature Communications 9 (2018). doi:10.1038/s41467-018-05953-4 (open access - download PDF). PDB IDs 6h0k, 6h0l, 6gw9 and 6gwa.
- S. Botha, D. Baitan, K. E. J. Jungnickel, D. Oberthür, et al. "De novoprotein structure determination by heavy-atom soaking in lipidic cubic phase and SIRAS phasing using serial synchrotron crystallography". IUCrJ 5 (2018) 524-530. doi:10.1107/s2052252518009223 (open access - download PDF). PDB ID 6fjs.
- Mary E O’Sullivan, Frédéric Poitevin, Raymond G Sierra, Cornelius Gati, et al. "Aminoglycoside ribosome interactions reveal novel conformational states at ambient temperature". Nucleic Acids Research 46 (2018) 9793-9804. doi:10.1093/nar/gky693 (open access - download PDF). PDB IDs 6cas and 6car.
- Hocheol Shin, Seungnam Kim and Chun Hong Yoon. "Data Analysis using Psocake at PAL-XFEL". Journal of the Korean Physical Society 73 (2018) 16-20. doi:10.3938/jkps.73.16
- Przemyslaw Nogly, Tobias Weinert, Daniel James, Sergio Carbajo, et al. "Retinal isomerization in bacteriorhodopsin captured by a femtosecond x-ray laser". Science 361 (2018). doi:10.1126/science.aat0094 PDB IDs 6g7h, 6g7i, 6g7j, 6g7k and 6g7l.
- Jose L. Olmos, Suraj Pandey, Jose M. Martin-Garcia, George Calvey, et al. "Enzyme intermediates captured “on the fly” by mix-and-inject serial crystallography". BMC Biology 16 (2018). doi:10.1186/s12915-018-0524-5 (open access - download PDF). PDB IDs 6b5x, 6b5y, 6b68, 6b69, 6b6a, 6b6b, 6b6c, 6b6d, 6b6e and 6b6f.
- Zhi Geng, Menglu Hu, Zhun She, Qiang Zhou, et al. "An iterative refinement method combining detector geometry optimization and diffraction model refinement in serial femtosecond crystallography". Radiation Detection Technology and Methods 2 (2018). doi:10.1007/s41605-017-0034-y
- Carrie L. Lomelino, Jin Kyun Kim, Cheol Lee, Seon Woo Lim, et al. "Carbonic anhydrase II microcrystals suitable for XFEL studies". Acta Crystallographica Section F Structural Biology Communications 74 (2018) 327-330. doi:10.1107/s2053230x18006118
- Dan Bi Lee, Jong-Min Kim, Jong Hyeon Seok, Ji-Hye Lee, et al. "Supersaturation-controlled microcrystallization and visualization analysis for serial femtosecond crystallography". Scientific Reports 8 (2018). doi:10.1038/s41598-018-20899-9 (open access - download PDF).
2017 (28 articles and 64 PDB entries this year)
- Robert Dods, Petra Båth, David Arnlund, Kenneth R. Beyerlein, et al. "From Macrocrystals to Microcrystals: A Strategy for Membrane Protein Serial Crystallography". Structure 25 (2017) 1461-1468.e2. doi:10.1016/j.str.2017.07.002 PDB IDs 5o4c, 5nj4 and 5o64.
- Xianjun Zhang, Fei Zhao, Yiran Wu, Jun Yang, et al. "Crystal structure of a multi-domain human smoothened receptor in complex with a super stabilizing ligand". Nature Communications 8 (2017). doi:10.1038/ncomms15383 (open access - download PDF). PDB ID 5v56.
- Nicolas Coquelle, Michel Sliwa, Joyce Woodhouse, Giorgio Schirò, et al. "Chromophore twisting in the excited state of a photoswitchable fluorescent protein captured by time-resolved serial femtosecond crystallography". Nature Chemistry 10 (2017) 31-37. doi:10.1038/nchem.2853 PDB IDs 5o8b, 5o8c, 5o8a and 5o89.
- Amit Sharma, Linda Johansson, Elin Dunevall, Weixiao Y. Wahlgren, et al. "Asymmetry in serial femtosecond crystallography data". Acta Crystallographica Section A Foundations and Advances 73 (2017) 93-101. doi:10.1107/s2053273316018696 (open access - download PDF). PDB IDs 5m7k and 5m7j.
- Ryoji Suno, Kanako Terakado Kimura, Takanori Nakane, Keitaro Yamashita, et al. "Crystal Structures of Human Orexin 2 Receptor Bound to the Subtype-Selective Antagonist EMPA". Structure 26 (2018) 7-19.e5. doi:10.1016/j.str.2017.11.005 PDB ID 5ws3. CXIDB ID 74.
- Takehiko Tosha, Takashi Nomura, Takuma Nishida, Naoya Saeki, et al. "Capturing an initial intermediate during the P450nor enzymatic reaction using time-resolved XFEL crystallography and caged-substrate". Nature Communications 8 (2017). doi:10.1038/s41467-017-01702-1 (open access - download PDF). PDB IDs 5y5i, 5y5j, 5y5k, 5y5l and 5y5m. CXIDB IDs 63 and 64.
- Thomas P. Halsted, Keitaro Yamashita, Kunio Hirata, Hideo Ago, et al. "An unprecedented dioxygen species revealed by serial femtosecond rotation crystallography in copper nitrite reductase". IUCrJ 5 (2018) 22-31. doi:10.1107/s2052252517016128 (open access - download PDF). PDB ID 5onx.
- Alexander Gorel, Koji Motomura, Hironobu Fukuzawa, R. Bruce Doak, et al. "Multi-wavelength anomalous diffraction de novo phasing using a two-colour X-ray free-electron laser with wide tunability". Nature Communications 8 (2017). doi:10.1038/s41467-017-00754-7 (open access - download PDF). PDB ID 5oer. CXIDB ID 66.
- Kenneth R. Beyerlein, Dennis Dierksmeyer, Valerio Mariani, Manuela Kuhn, et al. "Mix-and-diffuse serial synchrotron crystallography". IUCrJ 4 (2017) 769-777. doi:10.1107/s2052252517013124 (open access - download PDF). PDB IDs 5njp, 5njq, 5njr and 5njs.
- Tobias Weinert, Natacha Olieric, Robert Cheng, Steffen Brünle, et al. "Serial millisecond crystallography for routine room-temperature structure determination at synchrotrons". Nature Communications 8 (2017). doi:10.1038/s41467-017-00630-4 (open access - download PDF). PDB IDs 5njm, 5nlx, 5nqt, 5o5w, 5nm4 and 5nm5.
- Christopher Hutchison, Violeta Cordon-Preciado, Rhodri Morgan, Takanori Nakane, et al. "X-ray Free Electron Laser Determination of Crystal Structures of Dark and Light States of a Reversibly Photoswitching Fluorescent Protein at Room Temperature". International Journal of Molecular Sciences 18 (2017) 1918. doi:10.3390/ijms18091918 (open access - download PDF). PDB IDs 5ooz, 5oqe, 5oq9 and 5oqa.
- Minoru Kubo, Eriko Nango, Kensuke Tono, Tetsunari Kimura, et al. "Nanosecond pump–probe device for time-resolved serial femtosecond crystallography developed at SACLA". Journal of Synchrotron Radiation 24 (2017) 1086-1091. doi:10.1107/s160057751701030x (open access - download PDF). CXIDB ID 72.
- Keitaro Yamashita, Naoyuki Kuwabara, Takanori Nakane, Tomohiro Murai, et al. "Experimental phase determination with selenomethionine or mercury-derivatization in serial femtosecond crystallography". IUCrJ 4 (2017) 639-647. doi:10.1107/s2052252517008557 (open access - download PDF). PDB IDs 5xfc, 5xfd and 5xfe. CXIDB IDs 61 and 62.
- Kenneth R. Beyerlein, Thomas A. White, Oleksandr Yefanov, Cornelius Gati, et al. "FELIX: an algorithm for indexing multiple crystallites in X-ray free-electron laser snapshot diffraction images". Journal of Applied Crystallography 50 (2017) 1075-1083. doi:10.1107/s1600576717007506 (open access - download PDF).
- Hisashi Naitow, Yoshinori Matsuura, Kensuke Tono, Yasumasa Joti, et al. "Protein–ligand complex structure from serial femtosecond crystallography using soaked thermolysin microcrystals and comparison with structures from synchrotron radiation". Acta Crystallographica Section D Structural Biology 73 (2017) 702-709. doi:10.1107/s2059798317008919 (open access - download PDF). PDB IDs 5wr2, 5wr3 and 5wr4. CXIDB ID 73.
- X. Edward Zhou, Yuanzheng He, Parker W. de Waal, Xiang Gao, et al. "Identification of Phosphorylation Codes for Arrestin Recruitment by G Protein-Coupled Receptors". Cell 170 (2017) 457-469.e13. doi:10.1016/j.cell.2017.07.002 PDB ID 5w0p.
- Andrii Ishchenko, Daniel Wacker, Mili Kapoor, Ai Zhang, et al. "Structural insights into the extracellular recognition of the human serotonin 2B receptor by an antibody". Proceedings of the National Academy of Sciences 114 (2017) 8223-8228. doi:10.1073/pnas.1700891114 PDB ID 5tud.
- Izumi Ishigami, Nadia A. Zatsepin, Masahide Hikita, Chelsie E. Conrad, et al. "Crystal structure of CO-bound cytochrome c oxidase determined by serial femtosecond X-ray crystallography at room temperature". Proceedings of the National Academy of Sciences 114 (2017) 8011-8016. doi:10.1073/pnas.1705628114 PDB ID 5w97.
- Rebecka Andersson, Cecilia Safari, Robert Dods, Eriko Nango, et al. "Serial femtosecond crystallography structure of cytochrome c oxidase at room temperature". Scientific Reports 7 (2017). doi:10.1038/s41598-017-04817-z (open access - download PDF). PDB ID 5ndc.
- Atsuhiro Shimada, Minoru Kubo, Seiki Baba, Keitaro Yamashita, et al. "A nanosecond time-resolved XFEL analysis of structural changes associated with CO release from cytochrome c oxidase". Science Advances 3 (2017). doi:10.1126/sciadv.1603042
- Jose M. Martin-Garcia, Chelsie E. Conrad, Garrett Nelson, Natasha Stander, et al. "Serial millisecond crystallography of membrane and soluble protein microcrystals using synchrotron radiation". IUCrJ 4 (2017) 439-454. doi:10.1107/s205225251700570x (open access - download PDF). PDB IDs 5uvi, 5uvj, 5uvk and 5uvl.
- Haonan Zhang, Anna Qiao, Dehua Yang, Linlin Yang, et al. "Structure of the full-length glucagon class B G-protein-coupled receptor". Nature 546 (2017) 259-264. doi:10.1038/nature22363 PDB ID 5xez.
- Tetsuya Masuda, Mamoru Suzuki, Shigeyuki Inoue, Changyong Song, et al. "Atomic resolution structure of serine protease proteinase K at ambient temperature". Scientific Reports 7 (2017). doi:10.1038/srep45604 (open access - download PDF). PDB ID 5kxu. CXIDB ID 45.
- Haitao Zhang, Gye Won Han, Alexander Batyuk, Andrii Ishchenko, et al. "Structural basis for selectivity and diversity in angiotensin II receptors". Nature 544 (2017) 327-332. doi:10.1038/nature22035 PDB IDs 5unf, 5ung and 5unh.
- Michihiro Sugahara, Takanori Nakane, Tetsuya Masuda, Mamoru Suzuki, et al. "Hydroxyethyl cellulose matrix applied to serial crystallography". Scientific Reports 7 (2017). doi:10.1038/s41598-017-00761-0 (open access - download PDF). PDB IDs 5wr8, 5wr9, 5wra, 5wrb and 5wrc. CXIDB IDs 47, 48, 49 and 50.
- Dominik Oberthuer, Juraj Knoška, Max O. Wiedorn, Kenneth R. Beyerlein, et al. "Double-flow focused liquid injector for efficient serial femtosecond crystallography". Scientific Reports 7 (2017). doi:10.1038/srep44628 (open access - download PDF). PDB IDs 5u5q, 5mnd and 5trx.
- Michihiro Suga, Fusamichi Akita, Michihiro Sugahara, Minoru Kubo, et al. "Light-induced structural changes and the site of O=O bond formation in PSII caught by XFEL". Nature 543 (2017) 131-135. doi:10.1038/nature21400 PDB IDs 5ws5, 5ws6, 5gth and 5gti.
- Cornelius Gati, Dominik Oberthuer, Oleksandr Yefanov, Richard D. Bunker, et al. "Atomic structure of granulin determined from native nanocrystalline granulovirus using an X-ray free-electron laser". Proceedings of the National Academy of Sciences 114 (2017) 2247-2252. doi:10.1073/pnas.1609243114 PDB ID 5g0z.
2016 (21 articles and 46 PDB entries this year)
- Yohta Fukuda, Ka Man Tse, Mamoru Suzuki, Kay Diederichs, et al. "Redox-coupled structural changes in nitrite reductase revealed by serial femtosecond and microfocus crystallography". Journal of Biochemistry 159 (2016) 527-538. doi:10.1093/jb/mvv133 PDB ID 4ysa.
- Kazuya Hasegawa, Keitaro Yamashita, Tomohiro Murai, Nipawan Nuemket, et al. "Development of a dose-limiting data collection strategy for serial synchrotron rotation crystallography". Journal of Synchrotron Radiation 24 (2017) 29-41. doi:10.1107/s1600577516016362 (open access - download PDF). CXIDB ID 46.
- Eriko Nango, Antoine Royant, Minoru Kubo, Takanori Nakane, et al. "A three-dimensional movie of structural changes in bacteriorhodopsin". Science 354 (2016) 1552-1557. doi:10.1126/science.aah3497 PDB IDs 5b6v, 5b6w, 5h2h, 5h2i, 5h2j, 5b6x, 5h2k, 5h2l, 5h2m, 5b6y, 5h2n, 5h2o, 5h2p and 5b6z. CXIDB ID 53.
- Christopher Kupitz, Jose L. Olmos, Mark Holl, Lee Tremblay, et al. "Structural enzymology using X-ray free electron lasers". Structural Dynamics 4 (2016). doi:10.1063/1.4972069
- J. R. Stagno, Y. Liu, Y. R. Bhandari, C. E. Conrad, et al. "Structures of riboswitch RNA reaction states by mix-and-inject XFEL serial crystallography". Nature 541 (2016) 242-246. doi:10.1038/nature20599 PDB IDs 5e54, 5swd and 5swe.
- Mark S. Hunter, Chun Hong Yoon, Hasan DeMirci, Raymond G. Sierra, et al. "Selenium single-wavelength anomalous diffraction de novo phasing using an X-ray-free electron laser". Nature Communications 7 (2016). doi:10.1038/ncomms13388 (open access - download PDF). PDB ID 5jd2. CXIDB IDs 54 and 55.
- Takanori Nakane, Shinya Hanashima, Mamoru Suzuki, Haruka Saiki, et al. "Membrane protein structure determination by SAD, SIR, or SIRAS phasing in serial femtosecond crystallography using an iododetergent". Proceedings of the National Academy of Sciences 113 (2016) 13039-13044. doi:10.1073/pnas.1602531113 PDB IDs 5b34 and 5b35. CXIDB ID 34.
- Petra Edlund, Heikki Takala, Elin Claesson, Léocadie Henry, et al. "The room temperature crystal structure of a bacterial phytochrome determined by serial femtosecond crystallography". Scientific Reports 6 (2016). doi:10.1038/srep35279 (open access - download PDF). PDB IDs 5l8m and 5lbr. CXIDB ID 52.
- Alexander Batyuk, Lorenzo Galli, Andrii Ishchenko, Gye Won Han, et al. "Native phasing of x-ray free-electron laser data for a G protein–coupled receptor". Science Advances 2 (2016). doi:10.1126/sciadv.1600292 PDB IDs 5k2a, 5k2b, 5k2c and 5k2d.
- Przemyslaw Nogly, Valerie Panneels, Garrett Nelson, Cornelius Gati, et al. "Lipidic cubic phase injector is a viable crystal delivery system for time-resolved serial crystallography". Nature Communications 7 (2016). doi:10.1038/ncomms12314 (open access - download PDF). PDB ID 5j7a.
- Tao Zhang, Yuanxin Gu and Haifu Fan. "Effect of impurities and post-experimental purification in SAD phasing with serial femtosecond crystallography data". Acta Crystallographica Section D Structural Biology 72 (2016) 789-794. doi:10.1107/s205979831600646x
- Kanupriya Pande, Christopher D. M. Hutchison, Gerrit Groenhof, Andy Aquila, et al. "Femtosecond structural dynamics drives the trans/cis isomerization in photoactive yellow protein". Science 352 (2016) 725-729. doi:10.1126/science.aad5081 PDB IDs 5hd3, 5hdc, 5hdd, 5hds and 5hd5.
- Soichiro Tsujino and Takashi Tomizaki. "Ultrasonic acoustic levitation for fast frame rate X-ray protein crystallography at room temperature". Scientific Reports 6 (2016). doi:10.1038/srep25558 (open access - download PDF). PDB IDs 5fdj and 5fek.
- Takanori Nakane, Yasumasa Joti, Kensuke Tono, Makina Yabashi, et al. "Data processing pipeline for serial femtosecond crystallography at SACLA". Journal of Applied Crystallography 49 (2016) 1035-1041. doi:10.1107/s1600576716005720 (open access - download PDF).
- Michihiro Sugahara, Changyong Song, Mamoru Suzuki, Tetsuya Masuda, et al. "Oil-free hyaluronic acid matrix for serial femtosecond crystallography". Scientific Reports 6 (2016). doi:10.1038/srep24484 (open access - download PDF). PDB IDs 5b1d, 5b1e, 5b1f and 5b1g.
- Fumitaka Mafuné, Ken Miyajima, Kensuke Tono, Yoshihiro Takeda, et al. "Microcrystal delivery by pulsed liquid droplet for serial femtosecond crystallography". Acta Crystallographica Section D Structural Biology 72 (2016) 520-523. doi:10.1107/s2059798316001480 PDB ID 5dm9. CXIDB ID 35.
- Karol Nass, Anton Meinhart, Thomas R. M. Barends, Lutz Foucar, et al. "Protein structure determination by single-wavelength anomalous diffraction phasing of X-ray free-electron laser data". IUCrJ 3 (2016) 180-191. doi:10.1107/s2052252516002980 (open access - download PDF). PDB ID 5fgt.
- Yohta Fukuda, Ka Man Tse, Takanori Nakane, Toru Nakatsu, et al. "Redox-coupled proton transfer mechanism in nitrite reductase revealed by femtosecond crystallography". Proceedings of the National Academy of Sciences 113 (2016) 2928-2933. doi:10.1073/pnas.1517770113 PDB IDs 4ysc and 5d4i. CXIDB ID 34.
- Jacques-Philippe Colletier, Michel Sliwa, François-Xavier Gallat, Michihiro Sugahara, et al. "Serial Femtosecond Crystallography and Ultrafast Absorption Spectroscopy of the Photoswitchable Fluorescent Protein IrisFP". The Journal of Physical Chemistry Letters 7 (2016) 882-887. doi:10.1021/acs.jpclett.5b02789 PDB ID 5fvf.
- Kartik Ayyer, Oleksandr M. Yefanov, Dominik Oberthür, Shatabdi Roy-Chowdhury, et al. "Macromolecular diffractive imaging using imperfect crystals". Nature 530 (2016) 202-206. doi:10.1038/nature16949 PDB IDs 5e7c and 5e79.
- Arjen J. Jakobi, Daniel M. Passon, Kèvin Knoops, Francesco Stellato, et al. "In celluloserial crystallography of alcohol oxidase crystals inside yeast cells". IUCrJ 3 (2016) 88-95. doi:10.1107/s2052252515022927 (open access - download PDF).
2015 (24 articles and 39 PDB entries this year)
- R. Fromme, S. Roy-Chowdhury, S. Basu, C. Yoon, et al. "Light Harvesting Protein Phycocyanin in high resolution using a femtosecond X-Ray laser". Worldwide Protein Data Bank (2014). doi:10.2210/pdb4q70/pdb (no associated journal article). PDB ID 4q70.
- Sabine Botha, Karol Nass, Thomas R. M. Barends, Wolfgang Kabsch, et al. "Room-temperature serial crystallography at synchrotron X-ray sources using slowly flowing free-standing high-viscosity microstreams". Acta Crystallographica Section D Biological Crystallography 71 (2015) 387-397. doi:10.1107/s1399004714026327 PDB IDs 4rln and 4rlm.
- Dianfan Li, Phillip J. Stansfeld, Mark S. P. Sansom, Aaron Keogh, et al. "Ternary structure reveals mechanism of a membrane diacylglycerol kinase". Nature Communications 6 (2015). doi:10.1038/ncomms10140 (open access - download PDF). PDB ID 4uyo.
- Raymond G Sierra, Cornelius Gati, Hartawan Laksmono, E Han Dao, et al. "Concentric-flow electrokinetic injector enables serial crystallography of ribosome and photosystem II". Nature Methods 13 (2015) 59-62. doi:10.1038/nmeth.3667 PDB ID 5br8.
- Takanori Nakane, Changyong Song, Mamoru Suzuki, Eriko Nango, et al. "Native sulfur/chlorine SAD phasing for serial femtosecond crystallography". Acta Crystallographica Section D Biological Crystallography 71 (2015) 2519-2525. doi:10.1107/s139900471501857x (open access - download PDF). PDB ID 4yop. CXIDB ID 33.
- Tao Zhang, Deqiang Yao, Jiawei Wang, Yuanxin Gu, et al. "Serial crystallographic analysis of protein isomorphous replacement data from a mixture of native and derivative microcrystals". Acta Crystallographica Section D Biological Crystallography 71 (2015) 2513-2518. doi:10.1107/s139900471501603x
- Lorenzo Galli, Sang-Kil Son, Thomas R. M. Barends, Thomas A. White, et al. "Towards phasing using high X-ray intensity". IUCrJ 2 (2015) 627-634. doi:10.1107/s2052252515014049 (open access - download PDF). PDB IDs 5c6j, 5c6i and 5c6l.
- Keitaro Yamashita, Dongqing Pan, Tomohiko Okuda, Michihiro Sugahara, et al. "An isomorphous replacement method for efficient de novo phasing for serial femtosecond crystallography". Scientific Reports 5 (2015). doi:10.1038/srep14017 (open access - download PDF). PDB IDs 5d9b and 5d9c. CXIDB ID 31.
- Thomas R. M. Barends, Lutz Foucar, Albert Ardevol, Karol Nass, et al. "Direct observation of ultrafast collective motions in CO myoglobin upon ligand dissociation". Science 350 (2015) 445-450. doi:10.1126/science.aac5492 PDB IDs 5cmv, 5cn4, 5cn5, 5cn6, 5cn7, 5cn8, 5cn9, 5cnb, 5cnc, 5cnd, 5cne, 5cnf and 5cng.
- Raimund Fromme, Andrii Ishchenko, Markus Metz, Shatabdi Roy Chowdhury, et al. "Serial femtosecond crystallography of soluble proteins in lipidic cubic phase". IUCrJ 2 (2015) 545-551. doi:10.1107/s2052252515013160 (open access - download PDF). PDB IDs 4zix and 4ziz.
- Yanyong Kang, X. Edward Zhou, Xiang Gao, Yuanzheng He, et al. "Crystal structure of rhodopsin bound to arrestin by femtosecond X-ray laser". Nature 523 (2015) 561-567. doi:10.1038/nature14656 PDB ID 4zwj.
- Chelsie E. Conrad, Shibom Basu, Daniel James, Dingjie Wang, et al. "A novel inert crystal delivery medium for serial femtosecond crystallography". IUCrJ 2 (2015) 421-430. doi:10.1107/s2052252515009811 (open access - download PDF). PDB ID 4z8k.
- Maike Bublitz, Karol Nass, Nikolaj D. Drachmann, Anders J. Markvardsen, et al. "Structural studies of P-type ATPase–ligand complexes using an X-ray free-electron laser". IUCrJ 2 (2015) 409-420. doi:10.1107/s2052252515008969 (open access - download PDF). PDB ID 4xou.
- Marius Schmidt, Kanupriya Pande, Shibom Basu and Jason Tenboer. "Room temperature structures beyond 1.5 Å by serial femtosecond crystallography". Structural Dynamics 2 (2015). doi:10.1063/1.4919903
- L. Galli, S.-K. Son, M. Klinge, S. Bajt, et al. "Electronic damage in S atoms in a native protein crystal induced by an intense X-ray free-electron laser pulse". Structural Dynamics 2 (2015). doi:10.1063/1.4919398
- Nicolas Coquelle, Aaron S. Brewster, Ulrike Kapp, Anastasya Shilova, et al. "Raster-scanning serial protein crystallography using micro- and nano-focused synchrotron beams". Acta Crystallographica Section D Biological Crystallography 71 (2015) 1184-1196. doi:10.1107/s1399004715004514 (open access - download PDF). PDB IDs 4wg1, 4wg7, 4wl6 and 4wl7.
- Tao Zhang, Shifeng Jin, Yuanxin Gu, Yao He, et al. "SFX analysis of non-biological polycrystalline samples". IUCrJ 2 (2015) 322-326. doi:10.1107/s2052252515002146 (open access - download PDF).
- Haitao Zhang, Hamiyet Unal, Cornelius Gati, Gye Won Han, et al. "Structure of the Angiotensin Receptor Revealed by Serial Femtosecond Crystallography". Cell 161 (2015) 833-844. doi:10.1016/j.cell.2015.04.011 PDB ID 4yay. CXIDB ID 38.
- Thomas Barends, Thomas A. White, Anton Barty, Lutz Foucar, et al. "Effects of self-seeding and crystal post-selection on the quality of Monte Carlo-integrated SFX data". Journal of Synchrotron Radiation 22 (2015) 644-652. doi:10.1107/s1600577515005184
- Sébastien Boutet, Lutz Foucar, Thomas R. M. Barends, Sabine Botha, et al. "Characterization and use of the spent beam for serial operation of LCLS". Journal of Synchrotron Radiation 22 (2015) 634-643. doi:10.1107/s1600577515004002 (open access - download PDF). PDB IDs 4rw1 and 4rw2.
- Y. Feng, R. Alonso-Mori, T. R. M. Barends, V. D. Blank, et al. "Demonstration of simultaneous experiments using thin crystal multiplexing at the Linac Coherent Light Source". Journal of Synchrotron Radiation 22 (2015) 626-633. doi:10.1107/s1600577515003999 (open access - download PDF).
- Kensuke Tono, Eriko Nango, Michihiro Sugahara, Changyong Song, et al. "Diverse application platform for hard X-ray diffraction in SACLA (DAPHNIS): application to serial protein crystallography using an X-ray free-electron laser". Journal of Synchrotron Radiation 22 (2015) 532-537. doi:10.1107/s1600577515004464 (open access - download PDF). PDB ID 3wun.
- Yasumasa Joti, Takashi Kameshima, Mitsuhiro Yamaga, Takashi Sugimoto, et al. "Data acquisition system for X-ray free-electron laser experiments at SACLA". Journal of Synchrotron Radiation 22 (2015) 571-576. doi:10.1107/s1600577515004506 (open access - download PDF).
- Gustavo Fenalti, Nadia A Zatsepin, Cecilia Betti, Patrick Giguere, et al. "Structural basis for bifunctional peptide recognition at human δ-opioid receptor". Nature Structural & Molecular Biology 22 (2015) 265-268. doi:10.1038/nsmb.2965 PDB ID 4rwd. CXIDB ID 40.
- Przemyslaw Nogly, Daniel James, Dingjie Wang, Thomas A. White, et al. "Lipidic cubic phase serial millisecond crystallography using synchrotron radiation". IUCrJ 2 (2015) 168-176. doi:10.1107/s2052252514026487 (open access - download PDF). PDB ID 4x31.
2014 (17 articles and 16 PDB entries this year)
- Jason Tenboer, Shibom Basu, Nadia Zatsepin, Kanupriya Pande, et al. "Time-resolved serial crystallography captures high-resolution intermediates of photoactive yellow protein". Science 346 (2014) 1242-1246. doi:10.1126/science.1259357 PDB IDs 4wl9 and 4wla.
- Michihiro Sugahara, Eiichi Mizohata, Eriko Nango, Mamoru Suzuki, et al. "Grease matrix as a versatile carrier of proteins for serial crystallography". Nature Methods 12 (2014) 61-63. doi:10.1038/nmeth.3172 PDB IDs 3wul, 3wxt, 3wxu, 3wum, 4w4q, 3wxs and 3wxq.
- Michael R. Sawaya, Duilio Cascio, Mari Gingery, Jose Rodriguez, et al. "Protein crystal structure obtained at 2.9 Å resolution from injecting bacterial cells into an X-ray free-electron laser beam". Proceedings of the National Academy of Sciences 111 (2014) 12769-12774. doi:10.1073/pnas.1413456111 PDB IDs 4qx1 and 4qx3.
- Haiguang Liu and John C.H. Spence. "The indexing ambiguity in serial femtosecond crystallography (SFX) resolved using an expectation maximization algorithm". IUCrJ 1 (2014) 393-401. doi:10.1107/s2052252514020314 (open access - download PDF).
- Mark S. Hunter, Brent Segelke, Marc Messerschmidt, Garth J. Williams, et al. "Fixed-target protein serial microcrystallography with an x-ray free electron laser". Scientific Reports 4 (2014). doi:10.1038/srep06026 (open access - download PDF).
- Tao Zhang, Yang Li and Lijie Wu. "An alternative method for data analysis in serial femtosecond crystallography". Acta Crystallographica Section A Foundations and Advances 70 (2014) 670-676. doi:10.1107/s2053273314016982
- Christopher Kupitz, Shibom Basu, Ingo Grotjohann, Raimund Fromme, et al. "Serial time-resolved crystallography of photosystem II using a femtosecond X-ray laser". Nature 513 (2014) 261-265. doi:10.1038/nature13453 PDB IDs 4pbu and 4rvy.
- Martin Caffrey, Dianfan Li, Nicole Howe and Syed T. A. Shah. "‘Hit and run’ serial femtosecond crystallography of a membrane kinase in the lipid cubic phase". Philosophical Transactions of the Royal Society B: Biological Sciences 369 (2014) 20130621. doi:10.1098/rstb.2013.0621
- Oleksandr Yefanov, Cornelius Gati, Gleb Bourenkov, Richard A. Kirian, et al. "Mapping the continuous reciprocal space intensity distribution of X-ray serial crystallography". Philosophical Transactions of the Royal Society B: Biological Sciences 369 (2014) 20130333. doi:10.1098/rstb.2013.0333
- Thomas A. White. "Post-refinement method for snapshot serial crystallography". Philosophical Transactions of the Royal Society B: Biological Sciences 369 (2014) 20130330. doi:10.1098/rstb.2013.0330
- John C. H. Spence, Nadia A. Zatsepin and Chufeng Li. "Coherent convergent-beam time-resolved X-ray diffraction". Philosophical Transactions of the Royal Society B: Biological Sciences 369 (2014) 20130325. doi:10.1098/rstb.2013.0325
- Francesco Stellato, Dominik Oberthür, Mengning Liang, Richard Bean, et al. "Room-temperature macromolecular serial crystallography using synchrotron radiation". IUCrJ 1 (2014) 204-212. doi:10.1107/s2052252514010070 (open access - download PDF). PDB ID 4o34.
- Kun Qu, Liang Zhou and Yu-Hui Dong. "An improved integration method in serial femtosecond crystallography". Acta Crystallographica Section D Biological Crystallography 70 (2014) 1202-1211. doi:10.1107/s1399004714002004
- Matthias Frank, David B. Carlson, Mark S. Hunter, Garth J. Williams, et al. "Femtosecond X-ray diffraction from two-dimensional protein crystals". IUCrJ 1 (2014) 95-100. doi:10.1107/s2052252514001444 (open access - download PDF).
- Cornelius Gati, Gleb Bourenkov, Marco Klinge, Dirk Rehders, et al. "Serial crystallography onin vivogrown microcrystals using synchrotron radiation". IUCrJ 1 (2014) 87-94. doi:10.1107/s2052252513033939 (open access - download PDF). PDB ID 4n4z.
- Uwe Weierstall, Daniel James, Chong Wang, Thomas A. White, et al. "Lipidic cubic phase injector facilitates membrane protein serial femtosecond crystallography". Nature Communications 5 (2014). doi:10.1038/ncomms4309 PDB ID 4o9r. CXIDB ID 39.
- Changyong Song, Kensuke Tono, Jaehyun Park, Tomio Ebisu, et al. "Multiple application X-ray imaging chamber for single-shot diffraction experiments with femtosecond X-ray laser pulses". Journal of Applied Crystallography 47 (2014) 188-197. doi:10.1107/s1600576713029944
2013 (6 articles and 3 PDB entries this year)
- Wei Liu, Daniel Wacker, Cornelius Gati, Gye Won Han, et al. "Serial Femtosecond Crystallography of G Protein–Coupled Receptors". Science 342 (2013) 1521-1524. doi:10.1126/science.1244142 PDB ID 4nc3. CXIDB ID 21.
- Linda C. Johansson, David Arnlund, Gergely Katona, Thomas A. White, et al. "Structure of a photosynthetic reaction centre determined by serial femtosecond crystallography". Nature Communications 4 (2013). doi:10.1038/ncomms3911 (open access - download PDF). PDB ID 4cas.
- Thomas R. M. Barends, Lutz Foucar, Sabine Botha, R. Bruce Doak, et al. "De novo protein crystal structure determination from X-ray free-electron laser data". Nature 505 (2013) 244-247. doi:10.1038/nature12773 PDB ID 4n5r. CXIDB ID 20.
- Hasan Demirci, Raymond G. Sierra, Hartawan Laksmono, Robert L. Shoeman, et al. "Serial femtosecond X-ray diffraction of 30S ribosomal subunit microcrystals in liquid suspension at ambient temperature using an X-ray free-electron laser". Acta Crystallographica Section F Structural Biology and Crystallization Communications 69 (2013) 1066-1069. doi:10.1107/s174430911302099x (open access - download PDF).
- Thomas A. White, Anton Barty, Francesco Stellato, James M. Holton, et al. "Crystallographic data processing for free-electron laser sources". Acta Crystallographica Section D Biological Crystallography 69 (2013) 1231-1240. doi:10.1107/s0907444913013620 (open access - download PDF).
- Thomas R. M. Barends, Lutz Foucar, Robert L. Shoeman, Sadia Bari, et al. "Anomalous signal from S atoms in protein crystallographic data from an X-ray free-electron laser". Acta Crystallographica Section D Biological Crystallography 69 (2013) 838-842. doi:10.1107/s0907444913002448
2012 (5 articles and 4 PDB entries this year)
- Lars Redecke, Karol Nass, Daniel P. DePonte, Thomas A. White, et al. "Natively Inhibited Trypanosoma brucei Cathepsin B Structure Determined by Using an X-ray Laser". Science 339 (2013) 227-230. doi:10.1126/science.1229663 PDB ID 4hwy.
- Sébastien Boutet, Lukas Lomb, Garth J. Williams, Thomas R. M. Barends, et al. "High-Resolution Protein Structure Determination by Serial Femtosecond Crystallography". Science 337 (2012) 362-364. doi:10.1126/science.1217737 PDB IDs 4et8 and 4et9. CXIDB ID 16.
- Linda C Johansson, David Arnlund, Thomas A White, Gergely Katona, et al. "Lipidic phase membrane protein serial femtosecond crystallography". Nature Methods 9 (2012) 263-265. doi:10.1038/nmeth.1867 PDB ID 4ac5.
- Rudolf Koopmann, Karolina Cupelli, Lars Redecke, Karol Nass, et al. "In vivo protein crystallization opens new routes in structural biology". Nature Methods 9 (2012) 259-262. doi:10.1038/nmeth.1859
- Andrew Aquila, Mark S. Hunter, R. Bruce Doak, Richard A. Kirian, et al. "Time-resolved protein nanocrystallography using an X-ray free-electron laser". Optics Express 20 (2012) 2706. doi:10.1364/oe.20.002706
2011 (2 articles and 1 PDB entries this year)
- Anton Barty, Carl Caleman, Andrew Aquila, Nicusor Timneanu, et al. "Self-terminating diffraction gates femtosecond X-ray nanocrystallography measurements". Nature Photonics 6 (2011) 35-40. doi:10.1038/nphoton.2011.297
- Henry N. Chapman, Petra Fromme, Anton Barty, Thomas A. White, et al. "Femtosecond X-ray protein nanocrystallography". Nature 470 (2011) 73-77. doi:10.1038/nature09750 PDB ID 3pcq.


