 |
Millepede-II
V04-00-00_preview
|
Major changes with respect to the draft manual.
The following methods to obtain the solution 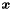 from a linear equation system
from a linear equation system 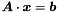 are implemented:
are implemented:
The solution and the covariance matrix 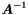 are obtained by inversion of
are obtained by inversion of 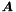 . Available are the value, error and global correlation for all global parameters. The matrix inversion routine has been parallelized and can be used for up to several 10000 parameters.
. Available are the value, error and global correlation for all global parameters. The matrix inversion routine has been parallelized and can be used for up to several 10000 parameters.
The solution and the covariance matrix 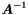 are obtained by diagonalization of
are obtained by diagonalization of 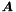 . Available are the value, error, global correlation and eigenvalue (and eigenvector) for all global parameters.
. Available are the value, error, global correlation and eigenvalue (and eigenvector) for all global parameters.
The solution is obtained by minimizing 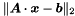 iteratively. MINRES is a special case of the generalized minimal residual method (GMRES) for symmetric matrices. Preconditioning with a band matrix of zero or finite bandwidth is possible. Individual columns
iteratively. MINRES is a special case of the generalized minimal residual method (GMRES) for symmetric matrices. Preconditioning with a band matrix of zero or finite bandwidth is possible. Individual columns 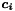 of the covariance matrix can be calculated by solving
of the covariance matrix can be calculated by solving 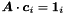 where
where 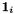 is the i-th column on the unit matrix. The most time consuming part (product matrix times vector per iteration) has been parallelized. Available are the value for all (and optionally error, global correlation for few) global parameters.
is the i-th column on the unit matrix. The most time consuming part (product matrix times vector per iteration) has been parallelized. Available are the value for all (and optionally error, global correlation for few) global parameters.
In case the local fit is a track fit with proper description of multiple scattering in the detector material additional local parameters have to be introduced for each scatterer and solution by inversion can get time consuming (~ 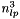 for
for 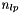 local parameters). For trajectories based on broken lines [ref 4,6] the corresponding matrix
local parameters). For trajectories based on broken lines [ref 4,6] the corresponding matrix 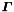 has a bordered band structure (
has a bordered band structure ( 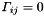 for
for 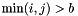 (border size) and
(border size) and 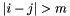 (bandwidth)). With root-free Cholesky decomposition the time for the solution is linear and for the calculation of
(bandwidth)). With root-free Cholesky decomposition the time for the solution is linear and for the calculation of 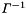 (needed for the construction of the global matrix) quadratic in
(needed for the construction of the global matrix) quadratic in 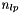 . For each local fit the structure of
. For each local fit the structure of 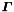 is checked and the faster solution method selected automatically.
is checked and the faster solution method selected automatically.
The code has been largely parallelized using OpenMP™. This includes the reading of binary files, the local fits, the construction of the sparsity structure and filling of the global matrix and the global fit (except by diagonalization). The number of threads is set by the command threads.
Caching. The records are read in blocks into a read cache and processed from there in parallel, each record by a single thread. For the filling of the global matrix the (zero-compressed) update matrices ( 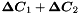 from equations (15), (16)) produced by each local fit are collected in a write cache. After processing the block of records this is used to update the global matrix in parallel, each row by a single thread. The total cache size can be changed by the command cache.
from equations (15), (16)) produced by each local fit are collected in a write cache. After processing the block of records this is used to update the global matrix in parallel, each row by a single thread. The total cache size can be changed by the command cache.
In sparse storage mode for each row the list of column indices (and values) for the non-zero elements are stored. With compression regions of continous column indices are represented by the first index and their number (packed into a single 32bit integer). Compression is selected by the command compress. In addition rare elements can be neglected (,histogrammed) or stored in single instead of double precision according to the pairentries command.
The zlib can be used to directly read gzipped C binary files. In this case reading with multiple threads (each file by single thread) can speed up the decompression.
The Millepede source code has been formally transformed from fixed form FORTRAN77 to free form Fortran90 (using TO_F90 by Alan Miller) and (most parts) modernized:
IMPLICIT NONE everywhere. Unused variables removed.COMMON blocks replaced by MODULEs.GOTOs replaced by proper DO loops.INTENT (input/output) of arguments described.Unused parts of the code (like the interactive mode) have been removed. The reference compiler for the Fortran90 version is gcc-4.6.2 (gcc-4.4.4 works too).
The memory management for dynamic data structures (matrices, vectors, ..) has been changed from a subdivided static COMMON block to dynamic (ALLOCATABLE) Fortran90 arrays. One Pede executable is now sufficient for all application sizes.
In the first loop over all binary files a preset read buffer size is used. Too large records are skipped, but the maximal record length is still being updated. If any records had to be skipped the read buffer size is afterwards adjusted according to the maximal record length and the first loop is repeated.
The number of binary files has no hard-coded limit anymore, but is calculated from the steering file and resources (file names, descriptors, ..) are allocated dynamically.
 1.8.1
1.8.1