URL: https://www.desy.de/news/news_search/index_eng.html
Breadcrumb Navigation
DESY News: X-ray pulses reveal structure of viral cocoon
News
News from the DESY research centre
X-ray pulses reveal structure of viral cocoon
An international team of scientists has used high-intensity X-ray pulses to determine the structure of the crystalline protein envelope of an insect virus. Their analysis reveals the fine details of the building blocks that make up the viral cocoon down to a scale of 0.2 nanometres (millionths of a millimetre) – approaching atom-scale resolution. The tiny viruses with their crystal casing are by far the smallest protein crystals ever analysed using X-ray crystallography. This opens up new opportunities in the study of protein structures, as the team headed by DESY’s Leading Scientist Henry Chapman from the Center for Free-Electron Laser Science reports in the Proceedings of the U.S. National Academy of Sciences (PNAS).
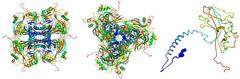
Atomic model of the crystalline occlusion bodies, derived from the X-ray diffraction images recorded at the LCLS. The individual proteins (right) stick together to form the building blocks (left, seen from the side; centre, seen from above) of the crystalline occlusion bodies. Credit: Dominik Oberthür, CFEL/DESY
Scientists are interested in the spatial structure of proteins and other biomolecules because this sheds light on the precise way in which they work. This has led to a specialised science known as structural biology. “Over the past 50 years, scientists have determined the structures of more than 100,000 proteins,” says Chapman, who is also a professor of physics at the University of Hamburg. “By far the most important tool for this is X-ray crystallography.” In this method, a crystal of the protein under investigation is grown and irradiated with bright X-rays. This produces a characteristic diffraction pattern, from which the spatial structure of the crystal and its building blocks can be calculated.

“These virus particles provided us with the smallest protein crystals ever used for X-ray structure analysis,” explains Gati. The occlusion body (the virus “cocoon”) has a volume of around 0.01 cubic micrometres, about one hundred times smaller than the smallest artificially grown protein crystals that have until now been analysed using crystallographic techniques.
To break this limit in crystal size, an extremely bright X-ray beam was needed, which was obtained using a so-called free-electron laser (FEL), in which a beam of high-speed electrons is guided through a magnetic undulator causing them to emit laser-like X-ray pulses.

Scanning electron microscope image showing the homogeneous size of the virus particles. Field of View is about 5 microns. Credit: Peter Metcalf, University of Auckland
The FEL dose was certainly lethal for the viruses too – each was completely vaporised by a single X-ray pulse. But the femtosecond-duration pulse carries the information of the pristine structure to the detector and the destruction of the virus occurs only after the passage of the pulse. The analysis of the recorded diffraction showed that even tiny protein crystals which are bombarded with extremely high radiation doses can still reveal their structure on an atomic scale.
“Simulations based on our measurements suggest that our method can probably be used to determine the structure of even smaller crystals consisting of only hundreds or thousands of molecules,” reports Chapman, who is also a member of the Hamburg Center for Ultrafast Imaging (CUI). “This takes us a huge step further towards our goal of analysing individual molecules.”
The European XFEL GmbH in Schenefeld, the Max Planck Institute for Medical Research in Heidelberg, the Arizona State University (USA) and the University of Basel (Switzerland) were also involved in the research.
Reference:
Atomic structure of granulin determined from native nanocrystalline granulovirus using an X-ray free-electron laser; Cornelius Gati et al.; PNAS, 2017; DOI: 10.1073/pnas.1609243114